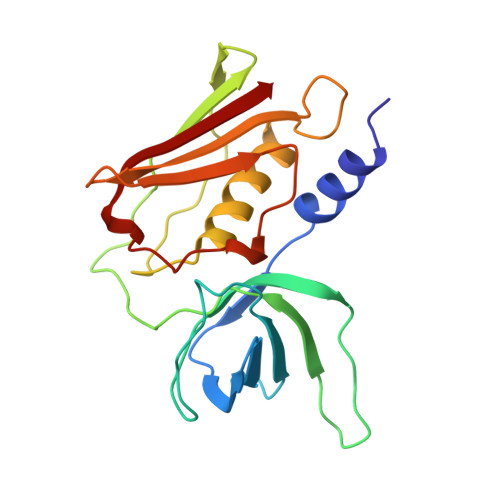
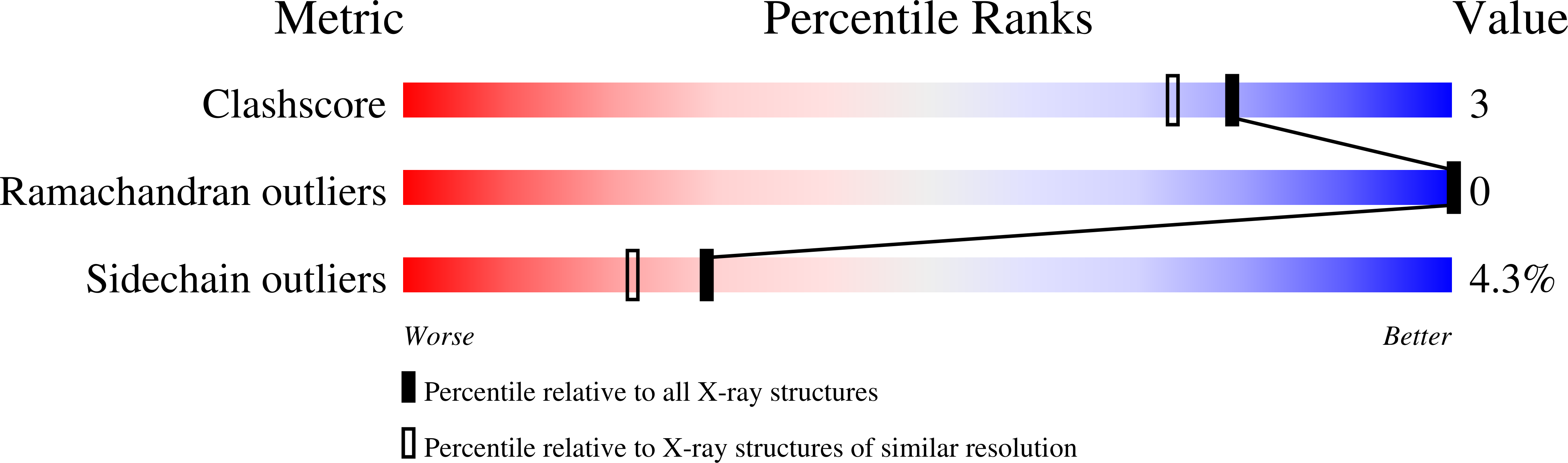The refined crystal structure of toxic shock syndrome toxin-1 at 2.07 A resolution.
Papageorgiou, A.C., Brehm, R.D., Leonidas, D.D., Tranter, H.S., Acharya, K.R.(1996) J Mol Biol 260: 553-569
- PubMed: 8759320
- DOI: https://doi.org/10.1006/jmbi.1996.0421
- Primary Citation of Related Structures:
2QIL - PubMed Abstract:
The pyrogenic toxin toxic shock syndrome toxin-1 from Staphylococcus aureus is a causative agent of the toxic shock syndrome disease. It belongs to a family of proteins known as superantigens that cross-link major histocompatibility class II molecules and T-cell receptors leading to the activation of a substantial number of T cells. The crystal structure of this protein has been refined to 2.07 A with an Rcryst value of 20.4% for 51,240 reflections. The final model contains three molecules in the asymmetric unit with good stereochemistry and a root-mean-square deviation of 0.009 A and 1.63 from ideality for bond lengths and bond angles, respectively. The overall fold is considerably similar to that of other known microbial superantigens (staphylococcal enterotoxins). However, a detailed structural analysis shows that toxic shock syndrome toxin-1 lacks several structural features that affect its specificity for V beta elements of the T-cell receptor and also its recognition by major histocompatibility class II molecules.
Organizational Affiliation:
School of Biology and Biochemistry, University of Bath, Claverton Down, UK.