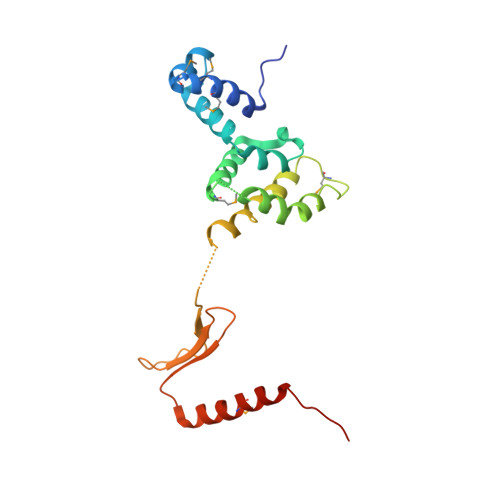
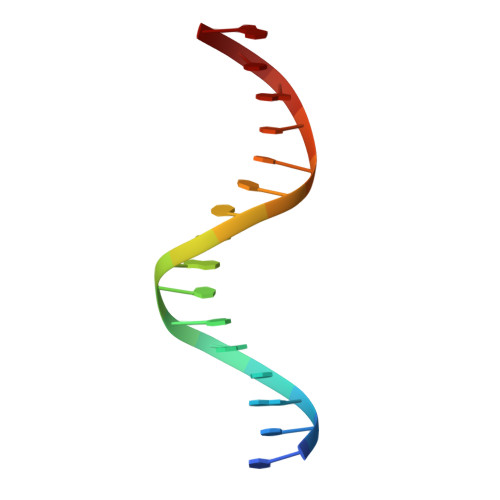
Structure of a four-way bridged ParB-DNA complex provides insight into P1 segrosome assembly.
Schumacher, M.A., Mansoor, A., Funnell, B.E.(2007) J Biol Chem 282: 10456-10464
- PubMed: 17293348
- DOI: https://doi.org/10.1074/jbc.M610603200
- Primary Citation of Related Structures:
2NTZ - PubMed Abstract:
The plasmid partition process is essential for plasmid propagation and is mediated by par systems, consisting of centromere-like sites and two proteins, ParA and ParB. In the first step of partition by the archetypical P1 system, ParB binds a complicated centromere-like site to form a large nucleoprotein segrosome. ParB is a dimeric DNA-binding protein that can bridge between both A-boxes and B-boxes located on the centromere. Its helix-turn-helix domains bind A-boxes and the dimer domain binds B-boxes. Binding of the first ParB dimer nucleates the remaining ParB molecules onto the centromere site, which somehow leads to the formation of a condensed segrosome superstructure. To further understand this unique DNA spreading capability of ParB, we crystallized and determined the structure of a 1:2 ParB-(142-333):A3-B2-box complex to 3.35A resolution. The structure reveals a remarkable four-way, protein-DNA bridged complex in which both ParB helix-turn-helix domains simultaneously bind adjacent A-boxes and the dimer domain bridges between two B-boxes. The multibridging capability and the novel dimer domain-B-box interaction, which juxtaposes the DNA sites close in space, suggests a mechanism for the formation of the wrapped solenoid-like segrosome superstructure. This multibridging capability of ParB is likely critical in its partition complex formation and pairing functions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Texas M. D. Anderson Cancer Center, Houston, Texas 77030, USA. maschuma@mdanderson