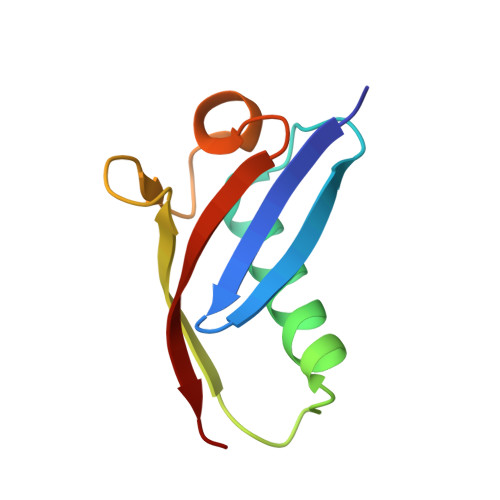
The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization
Feng, W., Wu, H., Chan, L.-N., Zhang, M.(2007) EMBO J 26: 2786-2796
- PubMed: 17476308
- DOI: https://doi.org/10.1038/sj.emboj.7601702
- Primary Citation of Related Structures:
2NS5 - PubMed Abstract:
The evolutionarily conserved Par-3/Par-6/aPKC complex is essential for the establishment and maintenance of polarity of a wide range of cells. Both Par-3 and Par-6 are PDZ domain containing scaffold proteins capable of binding to polarity regulatory proteins. In addition to three PDZ domains, Par-3 also contains a conserved N-terminal oligomerization domain (NTD) that is essential for proper subapical membrane localization and consequently the functions of Par-3. The molecular basis of NTD-mediated Par-3 membrane localization is poorly understood. Here, we describe the structure of a monomeric form of the Par-3 NTD. Unexpectedly, the domain adopts a PB1-like fold with both type-I and type-II structural features. The Par-3 NTD oligomerizes into helical filaments via front-to-back interactions. We further demonstrate that the NTD-mediated membrane localization of Par-3 in MDCK cells is solely attributed to its oligomerization capacity. The data presented in this study suggest that the Par-3 NTD is likely to facilitate the assembly of higher-order Par-3/Par-6/aPKC complex with increased avidities in targeting the complex to the subapical membrane domain and in binding to other polarity-regulating proteins.
Organizational Affiliation:
Department of Biochemistry, Molecular Neuroscience Center, The Hong Kong University of Science and Technology, Kowloon, Hong Kong.