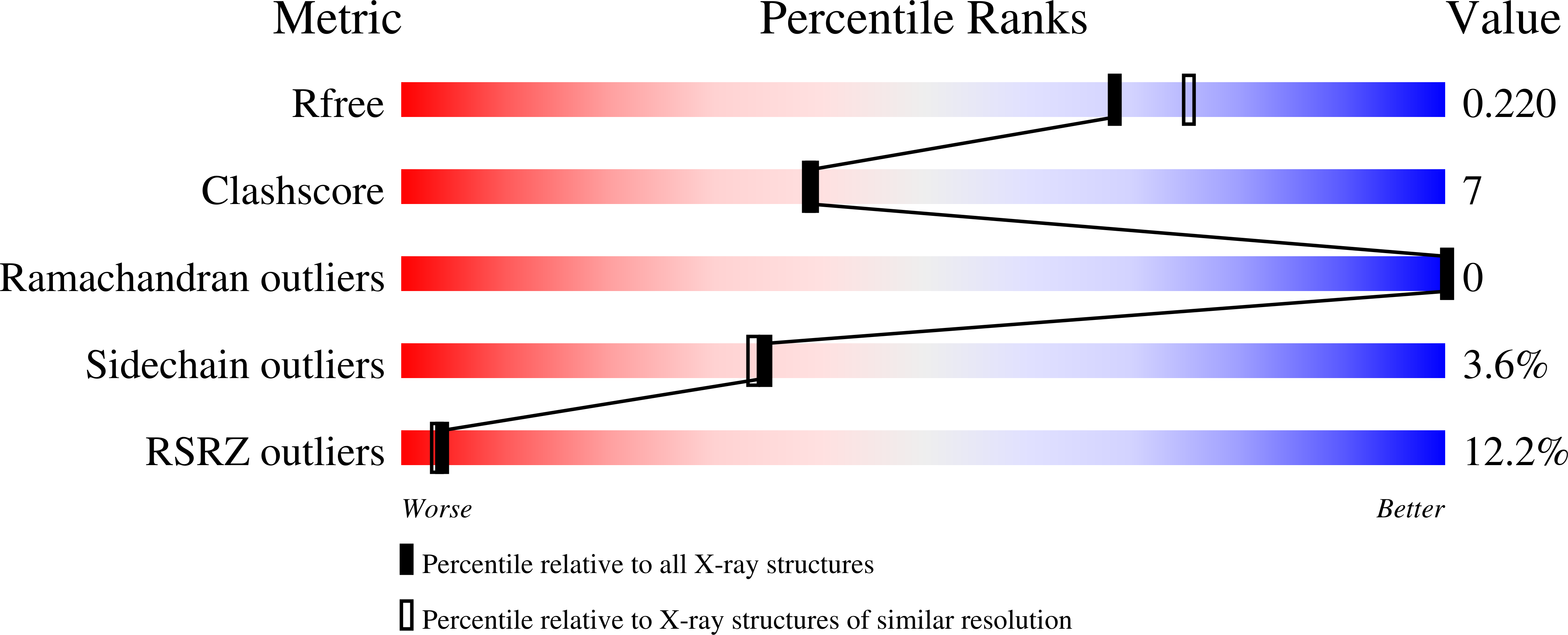
Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2).
Tang, Y., Lee, H.Y., Tang, Y., Kim, C.Y., Mathews, I., Khosla, C.(2006) Biochemistry 45: 14085-14093
- PubMed: 17115703
- DOI: https://doi.org/10.1021/bi061187v
- Primary Citation of Related Structures:
2NM0 - PubMed Abstract:
Aromatic polyketides are medicinally important natural products produced by type II polyketide synthases (PKSs). Some aromatic PKSs are bimodular and include a dedicated initiation module which synthesizes a non-acetate primer unit. Understanding the mechanism of this initiation module is expected to further enhance the potential for regiospecific modification of bacterial aromatic polyketides. A typical initiation module is comprised of a ketosynthase (KS), an acyl carrier protein (ACP), a malonyl-CoA:ACP transacylase (MAT), an acyl-ACP thioesterase, a ketoreductase (KR), a dehydratase (DH), and an enoyl reductase (ER). Thus far, the identities of the ketoreductase, dehydratase, and enoyl reductase remain a mystery because they are not encoded within the PKS biosynthetic gene cluster. Here we report that SCO1815 from Streptomyces coelicolor A3(2), an uncharacterized homologue of a NADPH-dependent ketoreductase, recognizes and reduces the beta-ketoacyl-ACP intermediate from the initiation module of the R1128 PKS. SCO1815 exhibits moderate specificity for both the acyl chain and the thiol donor. The X-ray crystal structure of SCO1815 was determined to 2.0 A. The structure shows that SCO1815 adopts a Rossmann fold and suggests that a conformational change occurs upon cofactor binding. We propose that a positively charged patch formed by three conserved residues is the ACP docking site. Our findings provide new engineering opportunities for incorporating unnatural primer units into novel polyketides and shed light on the biology of yet another cryptic protein in the S. coelicolor genome.
Organizational Affiliation:
Department of Chemistry, Stanford University, Stanford, California 94305, USA.