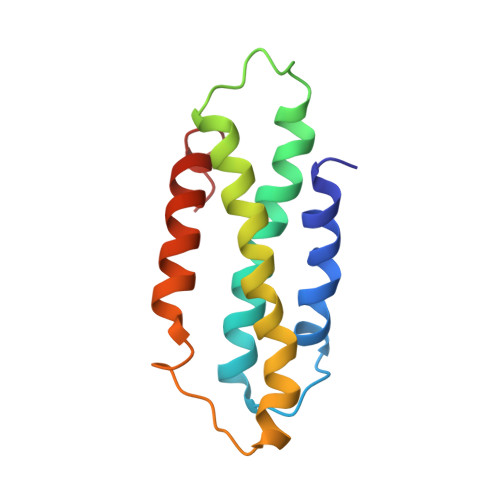
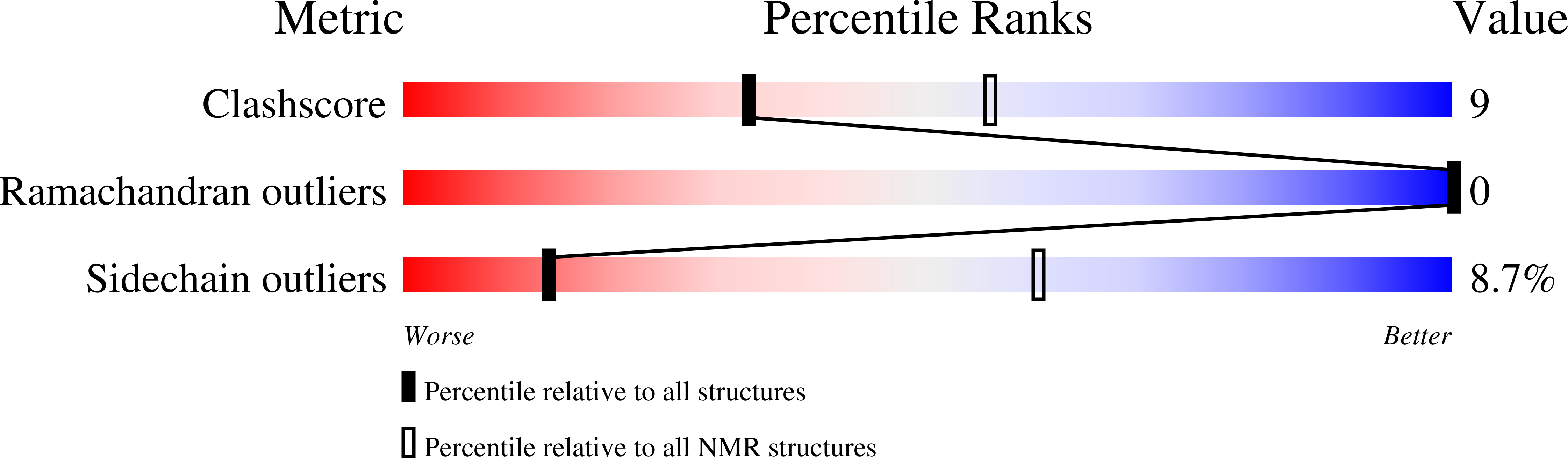The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins.
Casey, L.W., Lavrencic, P., Bentham, A.R., Cesari, S., Ericsson, D.J., Croll, T., Turk, D., Anderson, P.A., Mark, A.E., Dodds, P.N., Mobli, M., Kobe, B., Williams, S.J.(2016) Proc Natl Acad Sci U S A 113: 12856-12861
- PubMed: 27791121
- DOI: https://doi.org/10.1073/pnas.1609922113
- Primary Citation of Related Structures:
2NCG, 5T1Y - PubMed Abstract:
Plants use intracellular immunity receptors, known as nucleotide-binding oligomerization domain-like receptors (NLRs), to recognize specific pathogen effector proteins and induce immune responses. These proteins provide resistance to many of the world's most destructive plant pathogens, yet we have a limited understanding of the molecular mechanisms that lead to defense signaling. We examined the wheat NLR protein, Sr33, which is responsible for strain-specific resistance to the wheat stem rust pathogen, Puccinia graminis f. sp. tritici We present the solution structure of a coiled-coil (CC) fragment from Sr33, which adopts a four-helix bundle conformation. Unexpectedly, this structure differs from the published dimeric crystal structure of the equivalent region from the orthologous barley powdery mildew resistance protein, MLA10, but is similar to the structure of the distantly related potato NLR protein, Rx. We demonstrate that these regions are, in fact, largely monomeric and adopt similar folds in solution in all three proteins, suggesting that the CC domains from plant NLRs adopt a conserved fold. However, larger C-terminal fragments of Sr33 and MLA10 can self-associate both in vitro and in planta , and this self-association correlates with their cell death signaling activity. The minimal region of the CC domain required for both cell death signaling and self-association extends to amino acid 142, thus including 22 residues absent from previous biochemical and structural protein studies. These data suggest that self-association of the minimal CC domain is necessary for signaling but is likely to involve a different structural basis than previously suggested by the MLA10 crystallographic dimer.
Organizational Affiliation:
School of Chemistry and Molecular Biosciences, Institute for Molecular Bioscience and Australian Infectious Diseases Research Centre, University of Queensland, Brisbane, QLD 4072, Australia.