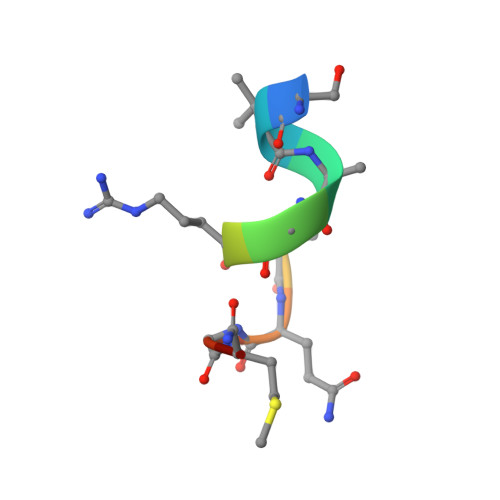
Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water.
Santana, M.J., de Oliveira, A.L., Queiroz Junior, L.H., Mandal, S.M., Matos, C.O., Dias, R.O., Franco, O.L., Liao, L.M.(2015) FEBS Lett 589: 639-644
- PubMed: 25639464
- DOI: https://doi.org/10.1016/j.febslet.2015.01.029
- Primary Citation of Related Structures:
2N0V - PubMed Abstract:
Multifunctional and promiscuous antimicrobial peptides (AMPs) can be used as an efficient strategy to control pathogens. However, little is known about the structural properties of plant promiscuous AMPs without disulfide bonds. CD and NMR were used to elucidate the structure of the promiscuous peptide Cn-AMP1, a disulfide-free peptide isolated from green coconut water. Data here reported shows that peptide structure is transitory and could be different according to the micro-environment. In this regard, Cn-AMP1 showed a random coil in a water environment and an α-helical structure in the presence of SDS-d25 micelles. Moreover, deuterium exchange experiments showed that Gly4, Arg5 and Met9 residues are less accessible to solvent, suggesting that flexibility and cationic charges seem to be essential for Cn-AMP1 multiple activities.
Organizational Affiliation:
Institute of Chemistry, Federal University of Goiás, Goiânia, GO, Brazil.