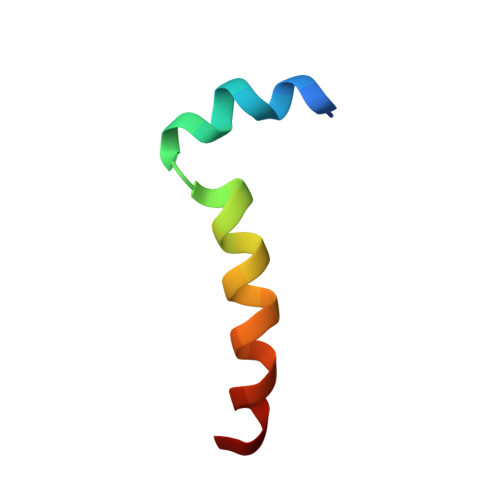
Thymosin alpha 1 inserts N terminus into model membranes assuming a helical conformation.
Nepravishta, R., Mandaliti, W., Eliseo, T., Sinibaldi Vallebona, P., Pica, F., Garaci, E., Paci, M.(2015) Expert Opin Biol Ther 15 Suppl 1: 71-81
- PubMed: 25642593
- DOI: https://doi.org/10.1517/14712598.2015.1009034
- Primary Citation of Related Structures:
2MNQ - PubMed Abstract:
Thymosin α1 (Tα1) is a peptide hormone whose therapeutic application has been approved in several diseases, but the description of a precise receptor for its therapeutic action still remains elusive and some knowledge of the mechanism of interaction with the cell membrane still needs to be clarified. This work is aimed at studying the folding and interaction of Tα1, which is completely unstructured in water solution, with model membranes. The folding and interaction of Tα1 with sodium dodecyl sulfate micelles was monitored by NMR and CD spectroscopy techniques. Tα1 assumes a helical conformation in the presence of sodium dodecyl sulfate micelles, showing a helical fold with a structural break around residues 9 and 14. These results were confirmed by circular dichroism and NMR spectroscopy. Moreover, by paramagnetic NMR relaxation it was found that Tα1 is inserted in the hydrophobic region of the micelles by the residues 1 - 5 of the N-terminal end. This result clarifies the modality of insertion that was not obtained in previous NMR studies in trifluoroethanol. These findings suggest that Tα1 folds on the membrane and, when inserted, may be able to interact with nearby proteins and/or receptors acting as an effector and causing a biological signaling cascade.
Organizational Affiliation:
University of Rome "Tor Vergata", Department of Chemical Sciences and Technologies , Via della Ricerca Scientifica 1, 00133 Rome , Italy paci@uniroma2.it.