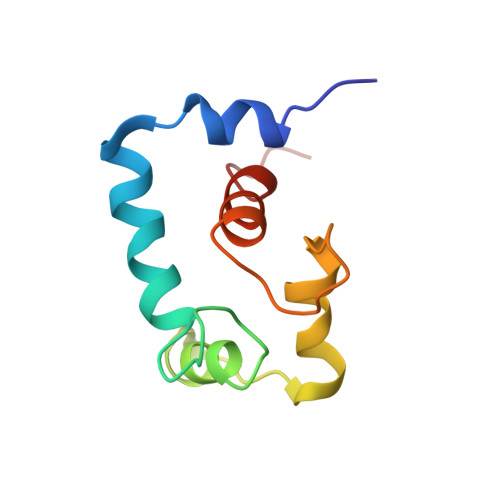
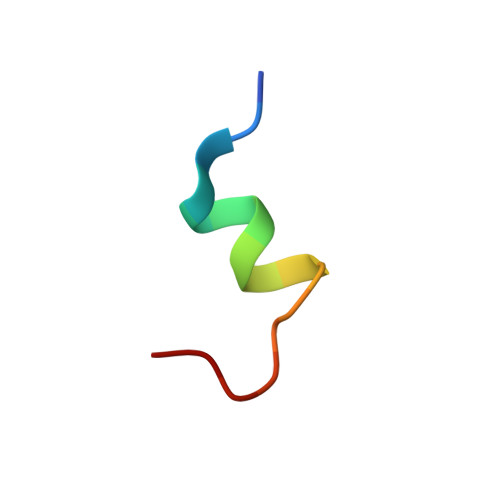
Conformation of the critical pH sensitive region of troponin depends upon a single residue in troponin I.
Robertson, I.M., Pineda-Sanabria, S.E., Holmes, P.C., Sykes, B.D.(2014) Arch Biochem Biophys 552-553: 40-49
- PubMed: 24333682
- DOI: https://doi.org/10.1016/j.abb.2013.12.003
- Primary Citation of Related Structures:
2MKP - PubMed Abstract:
The calcium sensitivity of cardiac and skeletal muscle is reduced during cytosolic acidosis, and this inhibition is more pronounced in cardiac muscle. Replacing cardiac troponin I with skeletal troponin I reduces the pH sensitivity of cardiac muscle. This diminished pH sensitivity depends on a single amino acid difference in troponin I: an alanine in cardiac and a histidine in skeletal. Studies suggested that when this histidine is protonated, it forms an electrostatic interaction with glutamate 19 on the surface of cardiac troponin C. Structures of the skeletal and cardiac troponin complexes show very different conformations for the region of troponin I surrounding this residue. In this study, we determined the structure of skeletal troponin I bound to cardiac troponin C. Skeletal troponin I is found to bind to cardiac troponin C with histidine 130 in close proximity to glutamate 19. This conformation is homologous to the crystal structure of the skeletal troponin complex; but different than in the cardiac complex. We show that an A162H variant of cardiac troponin I adopts a conformation similar to the skeletal structure. The implications of these structural differences in the context of cardiac muscle regulation are discussed.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, 4-19 Medical Sciences Building, Edmonton, Alberta T6G 2H7, Canada.