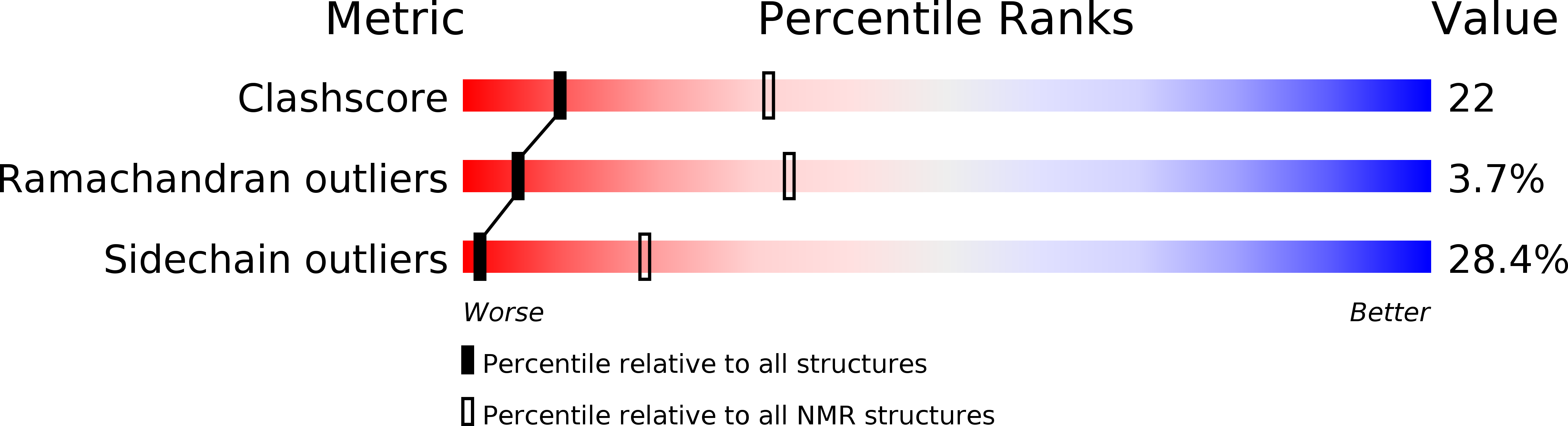Structural Basis for the Recognition of Cellular mRNA Export Factor REF by Herpes Viral Proteins HSV-1 ICP27 and HVS ORF57.
Tunnicliffe, R.B., Hautbergue, G.M., Kalra, P., Jackson, B.R., Whitehouse, A., Wilson, S.A., Golovanov, A.P.(2011) PLoS Pathog 7: e1001244-e1001244
- PubMed: 21253573
- DOI: https://doi.org/10.1371/journal.ppat.1001244
- Primary Citation of Related Structures:
2KT5 - PubMed Abstract:
The herpesvirus proteins HSV-1 ICP27 and HVS ORF57 promote viral mRNA export by utilizing the cellular mRNA export machinery. This function is triggered by binding to proteins of the transcription-export (TREX) complex, in particular to REF/Aly which directs viral mRNA to the TAP/NFX1 pathway and, subsequently, to the nuclear pore for export to the cytoplasm. Here we have determined the structure of the REF-ICP27 interaction interface at atomic-resolution and provided a detailed comparison of the binding interfaces between ICP27, ORF57 and REF using solution-state NMR. Despite the absence of any obvious sequence similarity, both viral proteins bind on the same site of the folded RRM domain of REF, via short but specific recognition sites. The regions of ICP27 and ORF57 involved in binding by REF have been mapped as residues 104-112 and 103-120, respectively. We have identified the pattern of residues critical for REF/Aly recognition, common to both ICP27 and ORF57. The importance of the key amino acid residues within these binding sites was confirmed by site-directed mutagenesis. The functional significance of the ORF57-REF/Aly interaction was also probed using an ex vivo cytoplasmic viral mRNA accumulation assay and this revealed that mutants that reduce the protein-protein interaction dramatically decrease the ability of ORF57 to mediate the nuclear export of intronless viral mRNA. Together these data precisely map amino acid residues responsible for the direct interactions between viral adaptors and cellular REF/Aly and provide the first molecular details of how herpes viruses access the cellular mRNA export pathway.
Organizational Affiliation:
Faculty of Life Sciences and Manchester Interdisciplinary Biocentre, University of Manchester, Manchester, United Kingdom.