Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
Mascioni, A., Bentley, B.E., Camarda, R., Dilts, D.A., Fink, P., Gusarova, V., Hoiseth, S.K., Jacob, J., Lin, S.L., Malakian, K., McNeil, L.K., Mininni, T., Moy, F., Murphy, E., Novikova, E., Sigethy, S., Wen, Y., Zlotnick, G.W., Tsao, D.H.(2009) J Biol Chem 284: 8738-8746
- PubMed: 19103601
- DOI: https://doi.org/10.1074/jbc.M808831200
- Primary Citation of Related Structures:
2KDY - PubMed Abstract:
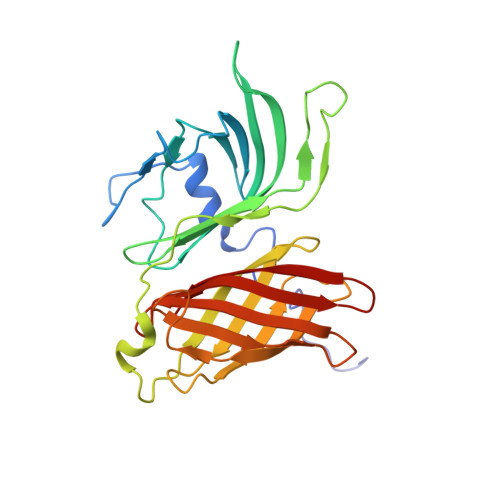
LP2086 is a family of outer membrane lipoproteins from Neisseria meningitidis, which elicits bactericidal antibodies and are currently undergoing human clinical trials in a bivalent formulation where each antigen represents one of the two known LP2086 subfamilies. Here we report the NMR structure of the recombinant LP2086 variant B01, a representative of the LP2086 subfamily B. The structure reveals a novel fold composed of two domains: a "taco-shaped" N-terminal beta-sheet and a C-terminal beta-barrel connected by a linker. The structure in micellar solution is consistent with a model of LP2086 anchored to the outer membrane bilayer through its lipidated N terminus. A long flexible chain connects the folded part of the protein to the lipid anchor and acts as spacer, making both domains accessible to the host immune system. Antibodies broadly reactive against members from both subfamilies have been mapped to the N terminus. A surface of subfamily-defining residues was identified on one face of the protein, offering an explanation for the induction of subfamily-specific bactericidal antibodies.
Organizational Affiliation:
Wyeth Research, Structural Biology and Computational Chemistry, Cambridge, Massachusetts 02140 and Wyeth Vaccines Research, Pearl River, New York 10965, USA.