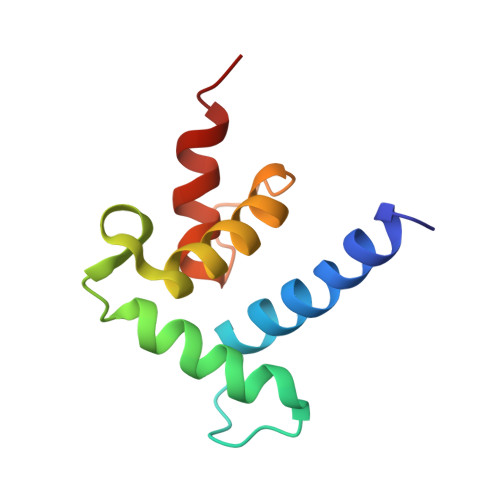
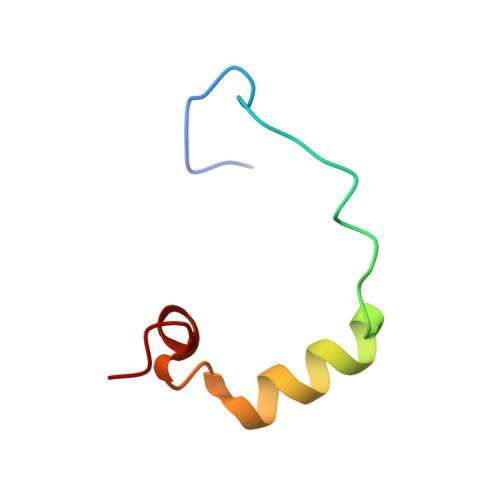
Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Wojciak, J.M., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E.(2009) EMBO J 28: 948-958
- PubMed: 19214187
- DOI: https://doi.org/10.1038/emboj.2009.30
- Primary Citation of Related Structures:
2KA4, 2KA6 - PubMed Abstract:
CBP/p300 transcriptional coactivators mediate gene expression by integrating cellular signals through interactions with multiple transcription factors. To elucidate the molecular and structural basis for CBP-dependent gene expression, we determined structures of the CBP TAZ1 and TAZ2 domains in complex with the transactivation domains (TADs) of signal transducer and activator of transcription 2 (STAT2) and STAT1, respectively. Despite the topological similarity of the TAZ1 and TAZ2 domains, subtle differences in helix packing and surface grooves constitute major determinants of target selectivity. Our results suggest that TAZ1 preferentially binds long TADs capable of contacting multiple surface grooves simultaneously, whereas smaller TADs that are restricted to a single contiguous binding surface form complexes with TAZ2. Complex formation for both STAT TADs involves coupled folding and binding, driven by intermolecular hydrophobic and electrostatic interactions. Phosphorylation of S727, required for maximal transcriptional activity of STAT1, does not enhance binding to any of the CBP domains. Because the different STAT TADs recognize different regions of CBP/p300, there is a potential for multivalent binding by STAT heterodimers that could enhance the recruitment of the coactivators to promoters.
Organizational Affiliation:
Department of Molecular Biology, The Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.