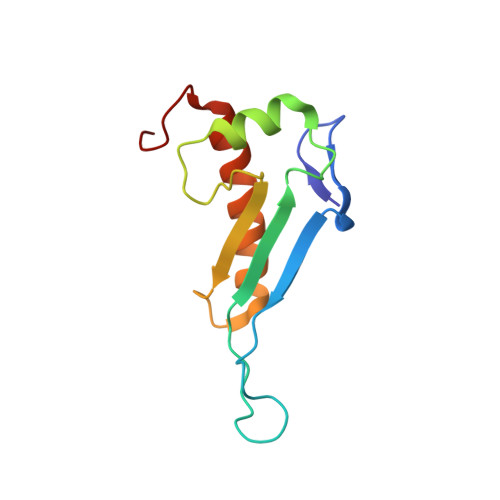
NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Singarapu, K.K., Xiao, R., Acton, T., Rost, B., Montelione, G.T., Szyperski, T.(2008) Proteins 71: 1027-1031
- PubMed: 18247350
- DOI: https://doi.org/10.1002/prot.21947
- Primary Citation of Related Structures:
2JVA - PubMed Abstract:
The NMR structure of the peptidyl-tRNA hydrolase (PTH) domain from Pseudomonas syringae ( P. syringae PTH domain) was solved by using recently devised protocol for high throughput protein structure determination. The P. syringae PTH domain belongs to a large Pfam family PF00472, which consists of at least 1549 proteins annotated as ‘hydrolysis domains of peptidyl-tRNA’. The structure of P. syringae PTH domain expands the ‘structural coverage’ of the PFam family.
Organizational Affiliation:
Department of Chemistry, State University of New York at Buffalo, Buffalo, New York 14260-3000, USA.