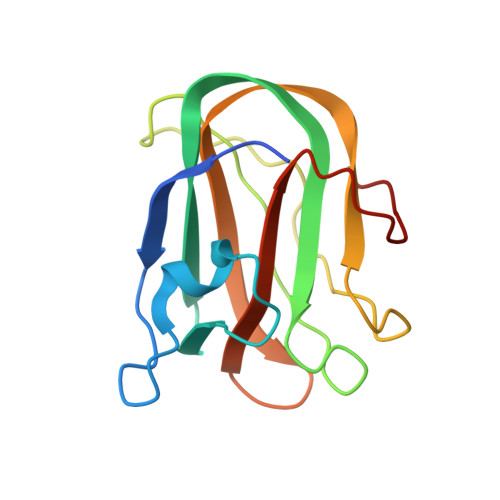
Identification and Characterization of a Novel Periplasmic Polygalacturonic Acid Binding Protein from Yersinia Enterolitica
Abbott, D.W., Hyrnuik, S., Boraston, A.B.(2007) J Mol Biol 367: 1023
- PubMed: 17292916
- DOI: https://doi.org/10.1016/j.jmb.2007.01.030
- Primary Citation of Related Structures:
2JD9, 2JDA - PubMed Abstract:
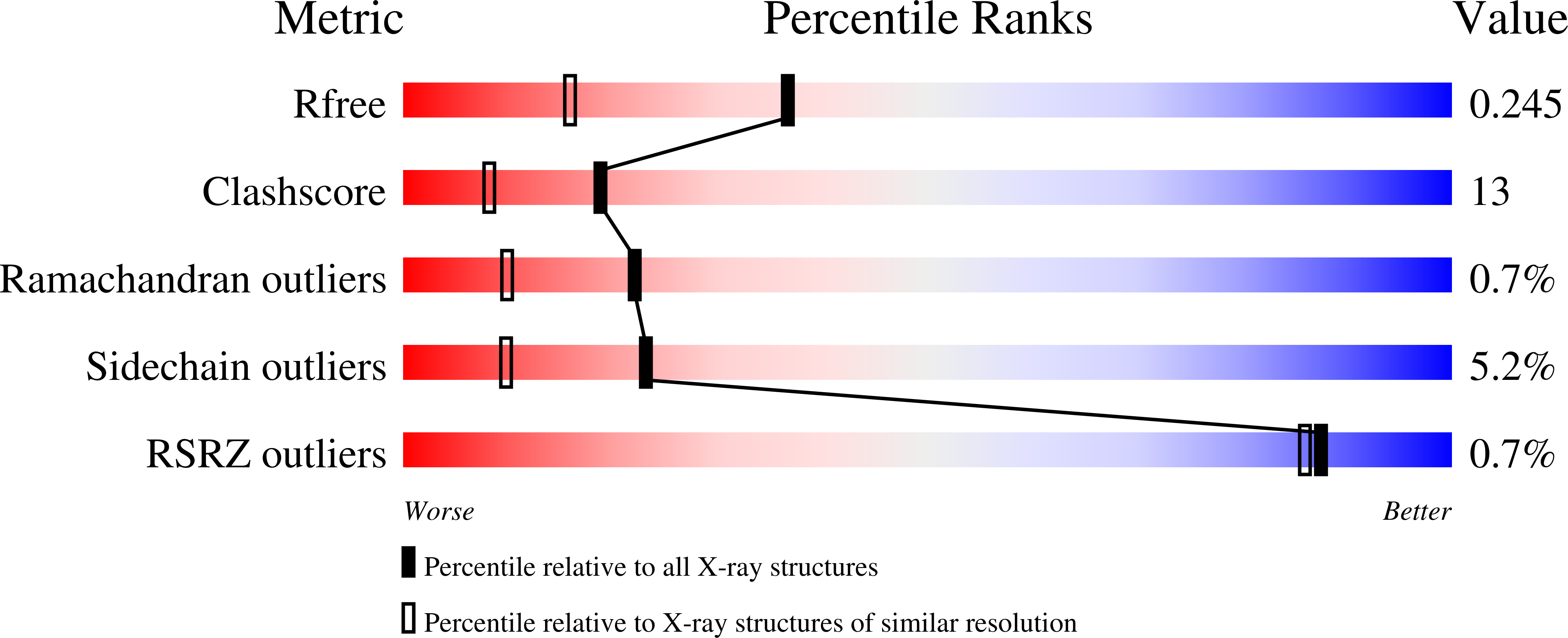
A number of bacteria in the family Enterobacteriaceae harbor the genes comprising well-developed pectinolytic pathways (e.g. Erwinia sp.) or abridged versions of this pathway (e.g. Yersinia sp.). One of the most enigmatic components present in some of these pathways is a small gene that encodes a predicted secreted protein of approximately 160 amino acid residues with unknown function. This protein shows distant amino acid sequence similarity over its entire length to galactose-specific family 32 carbohydrate-binding modules (CBMs). Here we demonstrate the ability of the Yersinia enterocolitica example, here called YeCBM32, to bind polygalacturonic acid containing components of pectin. This binding is selective for highly polymerized galacturonic acid and shows a complex mode of polysaccharide recognition. The high resolution X-ray crystal structure (1.35 A) shows YeCBM32s overall structural similarity to galactose specific CBMs and conserved binding site location but reveals a substantially different binding site topology, which likely reflects its unique polymeric and acidic ligand. The results suggest the possibility of a unique role for YeCBM32 in polygalacturonic acid transport.
Organizational Affiliation:
Biochemistry and Microbiology, University of Victoria, PO Box 3055 STN CSC, Victoria BC, Canada V8W 3P6.