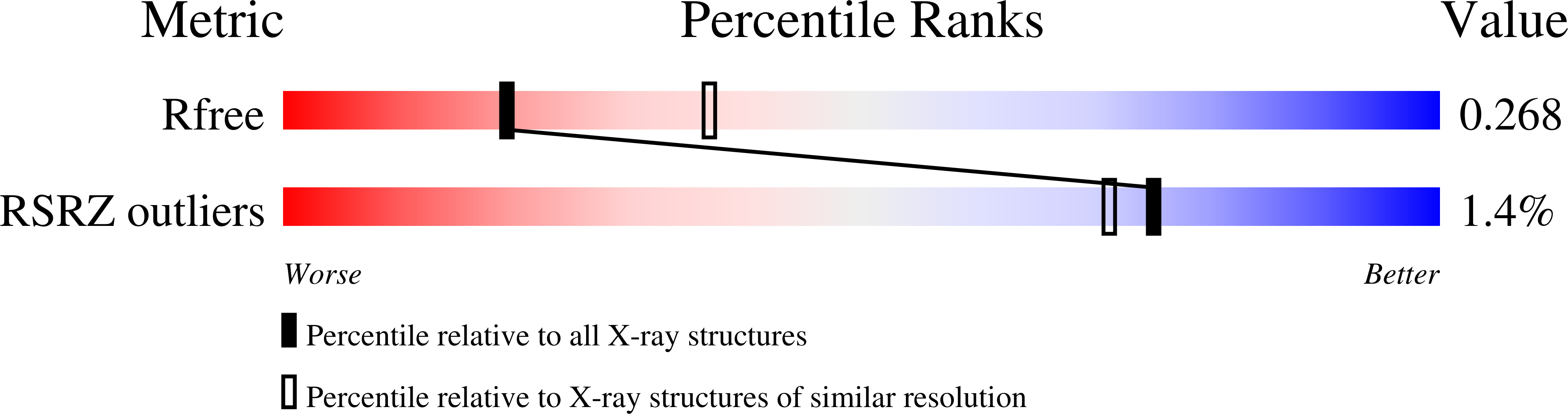Rationally Designed Small Compounds Inhibit Pilus Biogenesis in Uropathogenic Bacteria.
Pinkner, J.S., Remaut, H., Buelens, F., Miller, E., Aberg, V., Pemberton, N., Hedenstrom, M., Larsson, A., Seed, P., Waksman, G., Hultgren, S.J., Almqvist, F.(2006) Proc Natl Acad Sci U S A 103: 17897
- PubMed: 17098869
- DOI: https://doi.org/10.1073/pnas.0606795103
- Primary Citation of Related Structures:
2J7L - PubMed Abstract:
A chemical synthesis platform with broad applications and flexibility was rationally designed to inhibit biogenesis of adhesive pili assembled by the chaperone-usher pathway in Gram-negative pathogens. The activity of a family of bicyclic 2-pyridones, termed pilicides, was evaluated in two different pilus biogenesis systems in uropathogenic Escherichia coli. Hemagglutination mediated by either type 1 or P pili, adherence to bladder cells, and biofilm formation mediated by type 1 pili were all reduced by approximately 90% in laboratory and clinical E. coli strains. The structure of the pilicide bound to the P pilus chaperone PapD revealed that the pilicide bound to the surface of the chaperone known to interact with the usher, the outer-membrane assembly platform where pili are assembled. Point mutations in the pilicide-binding site dramatically reduced pilus formation but did not block the ability of PapD to bind subunits and mediate their folding. Surface plasmon resonance experiments confirmed that the pilicide interfered with the binding of chaperone-subunit complexes to the usher. These pilicides thus target key virulence factors in pathogenic bacteria and represent a promising proof of concept for developing drugs that function by targeting virulence factors.
Organizational Affiliation:
Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO 63110, USA.