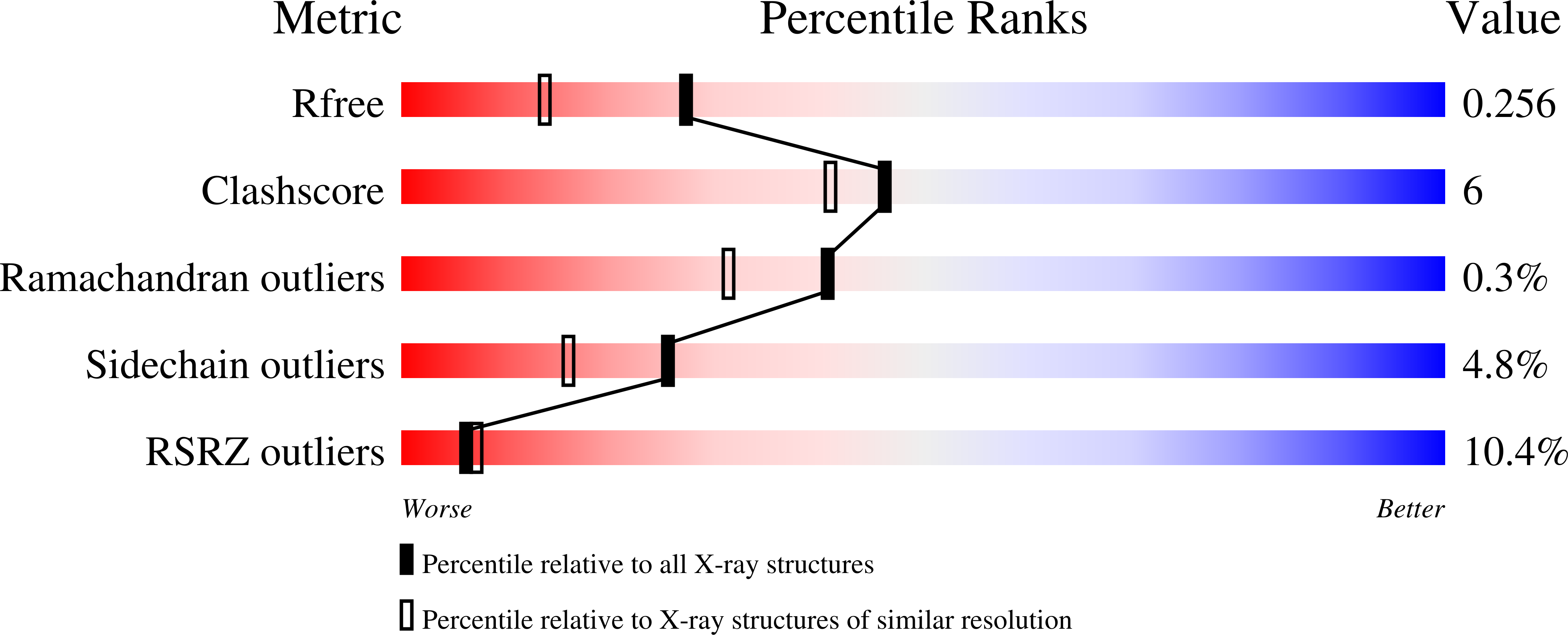
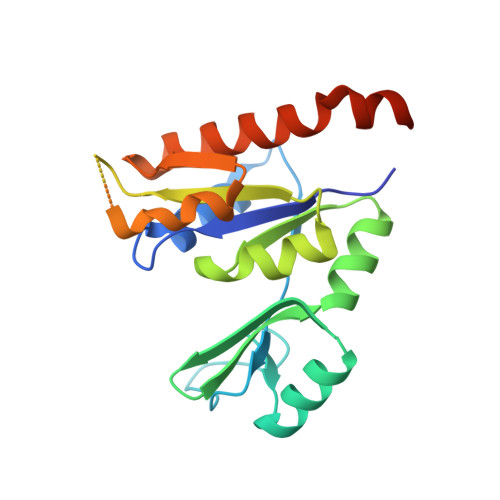
Structure of Staphylococcus Aureus Guanylate Monophosphate Kinase
El Omari, K., Dhaliwal, B., Lockyer, M., Charles, I., Hawkins, A.R., Stammers, D.K.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 949
- PubMed: 17012781
- DOI: https://doi.org/10.1107/S174430910603613X
- Primary Citation of Related Structures:
2J41 - PubMed Abstract:
Nucleotide monophosphate kinases (NMPKs) are potential antimicrobial drug targets owing to their role in supplying DNA and RNA precursors. The present work reports the crystal structure of Staphylococcus aureus guanylate monophosphate kinase (SaGMK) at 1.9 A resolution. The structure shows that unlike most GMKs SaGMK is dimeric, confirming the role of the extended C-terminus in dimer formation as first observed for Escherichia coli GMK (EcGMK). One of the two SaGMK dimers within the crystal asymmetric unit has two monomers in different conformations: an open form with a bound sulfate ion (mimicking the beta-phosphate of ATP) and a closed form with bound GMP and sulfate ion. GMP-induced domain movements in SaGMK can thus be defined by comparison of these conformational states. Like other GMKs, the binding of GMP firstly triggers a partial closure of the enzyme, diminishing the distance between the GMP-binding and ATP-binding sites. In addition, the closed structure shows the presence of a potassium ion in contact with the guanine ring of GMP. The potassium ion appears to form an integral part of the GMP-binding site, as the Tyr36 side chain has significantly moved to form a metal ion-ligand coordination involving the lone pair of the side-chain O atom. The potassium-binding site might also be exploited in the design of novel inhibitors.
Organizational Affiliation:
Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, England.