Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Massey, T.H., Mercogliano, C.P., Yates, J., Sherratt, D.J., Lowe, J.(2006) Mol Cell 23: 457
- PubMed: 16916635
- DOI: https://doi.org/10.1016/j.molcel.2006.06.019
- Primary Citation of Related Structures:
2IUS, 2IUT, 2IUU - PubMed Abstract:
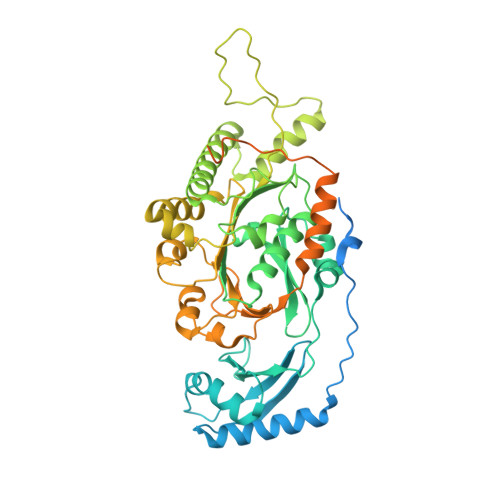
FtsK is a DNA translocase that coordinates chromosome segregation and cell division in bacteria. In addition to its role as activator of XerCD site-specific recombination, FtsK can translocate double-stranded DNA (dsDNA) rapidly and directionally and reverse direction. We present crystal structures of the FtsK motor domain monomer, showing that it has a RecA-like core, the FtsK hexamer, and also showing that it is a ring with a large central annulus and a dodecamer consisting of two hexamers, head to head. Electron microscopy (EM) demonstrates the DNA-dependent existence of hexamers in solution and shows that duplex DNA passes through the middle of each ring. Comparison of FtsK monomer structures from two different crystal forms highlights a conformational change that we propose is the structural basis for a rotary inchworm mechanism of DNA translocation.
Organizational Affiliation:
Division of Molecular Genetics, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, United Kingdom.