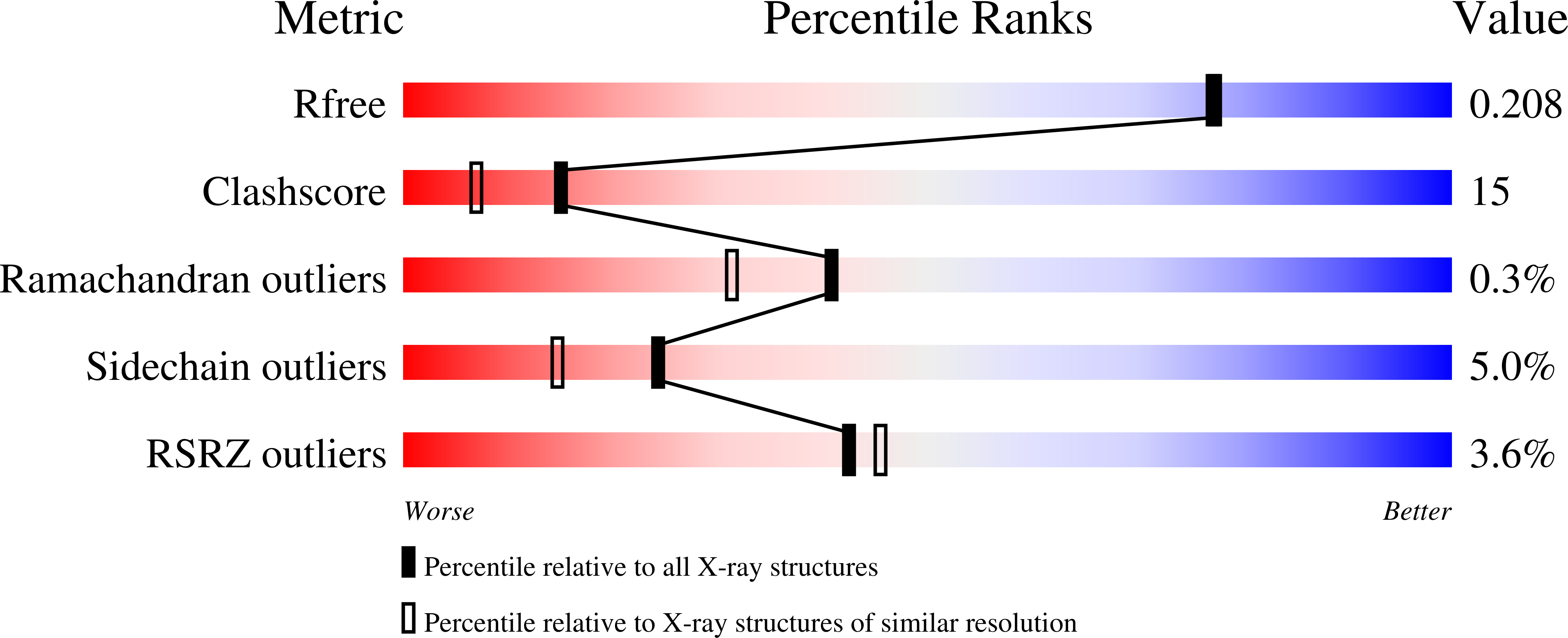
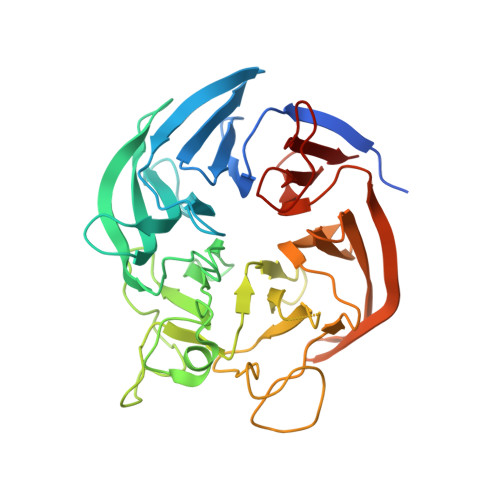
Crystal Structure of the Putative Oxidoreductase (Glucose Dehydrogenase) (TTHA0570) from Thermus Theromophilus HB8
Jeyakanthan, J., Kanaujia, S.P., Vasuki Ranjani, C., Sekar, K., Ebihara, A., Shinkai, A., Nakagawa, N., Shimizu, N., Yamamoto, M., Kuramitsu, S., Shiro, Y., Yokoyama, S.To be published.