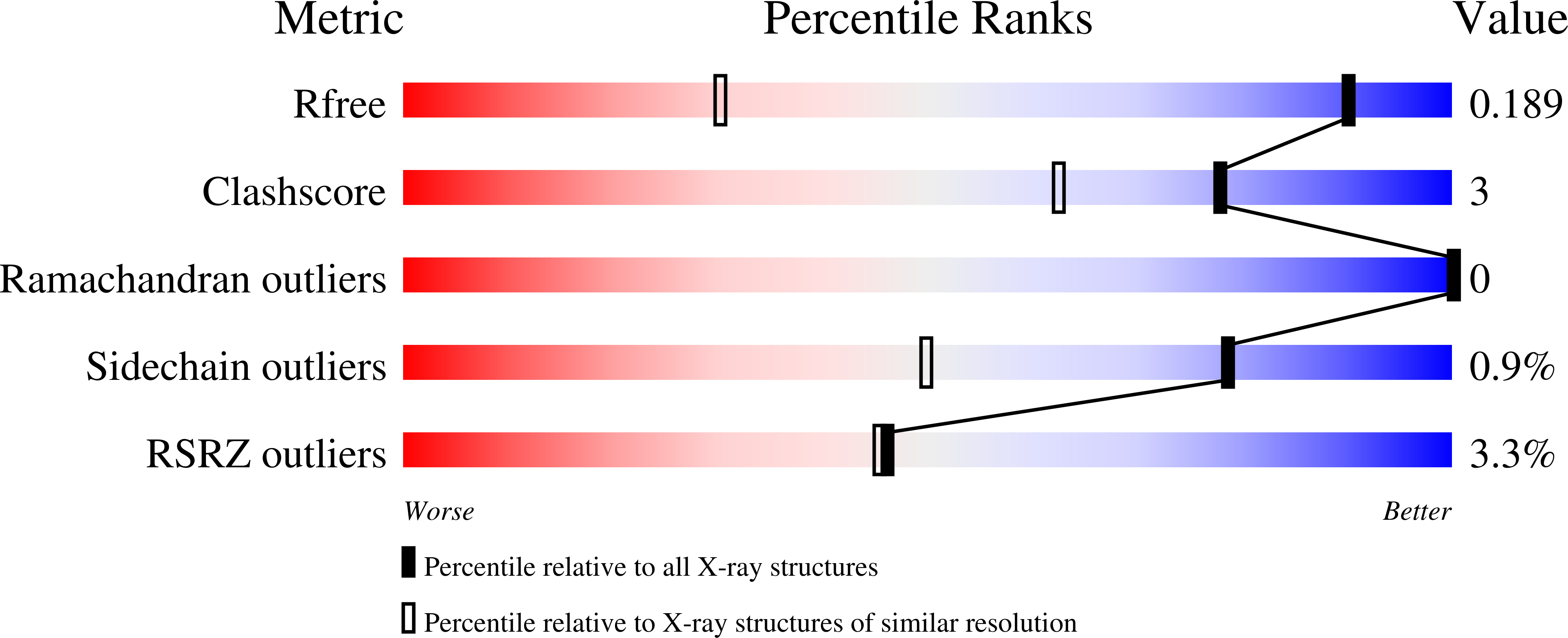
Hydrophobic surface burial is the major stability determinant of a flat, single-layer beta-sheet.
Yan, S., Gawlak, G., Makabe, K., Tereshko, V., Koide, A., Koide, S.(2007) J Mol Biol 368: 230-243
- PubMed: 17335845
- DOI: https://doi.org/10.1016/j.jmb.2007.02.003
- Primary Citation of Related Structures:
2I5Z - PubMed Abstract:
Formation of a flat beta-sheet is a fundamental event in beta-sheet-mediated protein self-assembly. To investigate the contributions of various factors to the stability of flat beta-sheets, we performed extensive alanine-scanning mutagenesis experiments on the single-layer beta-sheet segment of Borrelia outer surface protein A (OspA). This beta-sheet segment consists of beta-strands with highly regular geometries that can serve as a building block for self-assembly. Our Ala-scanning approach is distinct from the conventional host-guest method, in that it introduces only conservative, truncation mutations that should minimize structural perturbation. Our results showed very weak correlation with experimental beta-sheet propensity scales, statistical beta-sheet propensity scales, or cross-strand pairwise correlations. In contrast, our data showed strong positive correlation with the change in buried non-polar surface area. Polar interactions including prominent Glu-Lys cross-strand pairs contribute marginally to the beta-sheet stability. These results were corroborated by results from additional non-Ala mutations. Taken together, these results demonstrate the dominant contribution of non-polar surface burial to flat beta-sheet stability even at solvent-exposed positions. The OspA single-layer beta-sheet achieves efficient hydrophobic surface burial without forming a hydrophobic core by a strategic placement of a variety of side-chains. These findings further suggest the importance of hydrophobic interactions within a beta-sheet layer in peptide self-assembly.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.