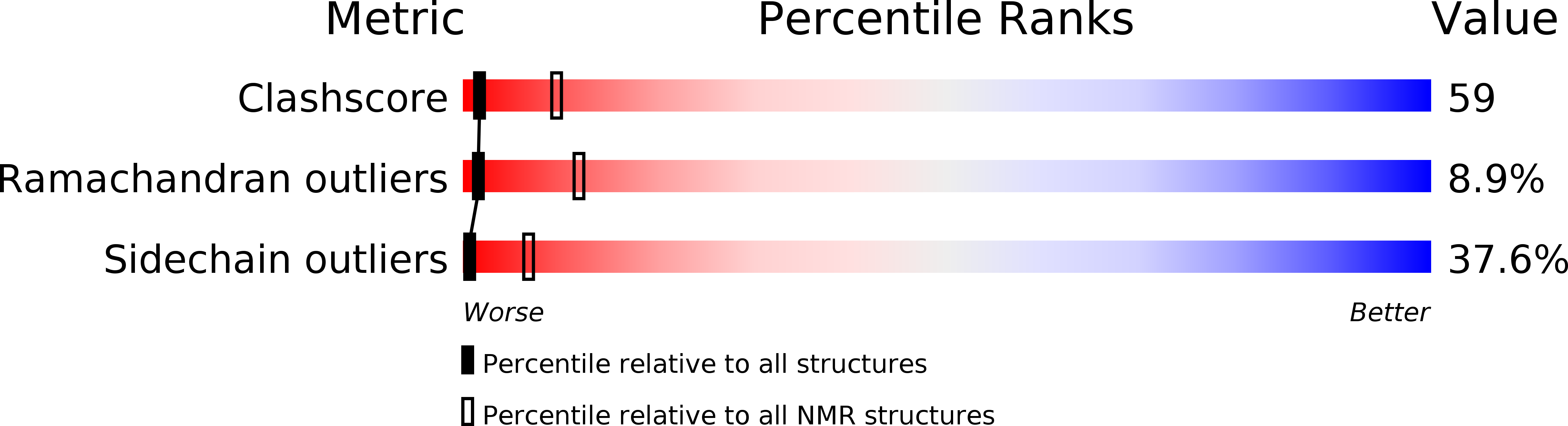Structural basis for the observed differential magnetic anisotropic tensorial values in calcium binding proteins.
Mustafi, S.M., Mukherjee, S., Chary, K.V., Cavallaro, G.(2006) Proteins 65: 656-669
- PubMed: 16981203
- DOI: https://doi.org/10.1002/prot.21121
- Primary Citation of Related Structures:
2I18 - PubMed Abstract:
Lanthanide ions (Ln(3+)), which have ionic radii similar to those of Ca(2+), can displace the latter in a calcium binding protein, without affecting its tertiary structure. The paramagnetic Ln(3+) possesses large anisotropic magnetic susceptibilities and produce pseudocontact shifts (PCSs), which have r(-3) dependence. The PCS can be seen for spins as far as 45 A from the paramagnetic ion. They aid in structure refinement of proteins by providing long-range distance constraints. Besides, they can be used to determine the interdomain orientation in multidomain proteins. This is particularly important in the context of a calcium binding protein from Entamoeba histolytica (EhCaBP), which consists of two globular domains connected by a flexible linker region containing 8 residues. As a first step to obtain the interdomain orientation in EhCaBP, a suite of 2D and 3D heteronuclear experiments were recorded on EhCaBP by displacing calcium with Ce(3+), Ho(3+), Er(3+), Tm(3+), Dy(3+), and Yb(3+) ions in separate experiments, and the PCS of (1)H(N) and (15)N spins were measured. Such data have been used in the refinement of the individual domain structures of the protein in parallel with the calculation of the respective magnetic anisotropy tensorial values, which differ substantially (2.1-2.8 times) from what is found in other Ca(2+) binding loops. This study provides a structural basis for such variations in the magnetic anisotropy tensorial values.
Organizational Affiliation:
Department of Chemical Sciences, Tata Institute of Fundamental Research, Mumbai 400005, India.