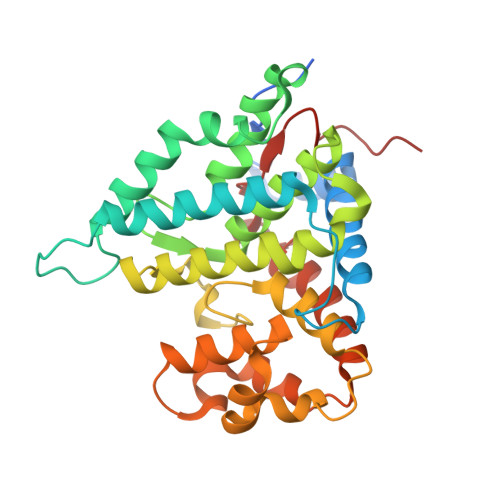
Structure and mechanism of the phosphotyrosyl phosphatase activator.
Chao, Y., Xing, Y., Chen, Y., Xu, Y., Lin, Z., Li, Z., Jeffrey, P.D., Stock, J.B., Shi, Y.(2006) Mol Cell 23: 535-546
- PubMed: 16916641
- DOI: https://doi.org/10.1016/j.molcel.2006.07.027
- Primary Citation of Related Structures:
2HV6, 2HV7 - PubMed Abstract:
Phosphotyrosyl phosphatase activator (PTPA), also known as PP2A phosphatase activator, is a conserved protein from yeast to human. Here we report the 1.9 A crystal structure of human PTPA, which reveals a previously unreported fold consisting of three subdomains: core, lid, and linker. Structural analysis uncovers a highly conserved surface patch, which borders the three subdomains, and an associated deep pocket located between the core and the linker subdomains. The conserved surface patch and the deep pocket are responsible for binding to PP2A and ATP, respectively. PTPA and PP2A A-C dimer together constitute a composite ATPase. PTPA binding to PP2A results in a dramatic alteration of substrate specificity, with enhanced phosphotyrosine phosphatase activity and decreased phosphoserine phosphatase activity. This function of PTPA strictly depends on the composite ATPase activity. These observations reveal significant insights into the function and mechanism of PTPA and have important ramifications for understanding PP2A function.
Organizational Affiliation:
Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, New Jersey 08544, USA.