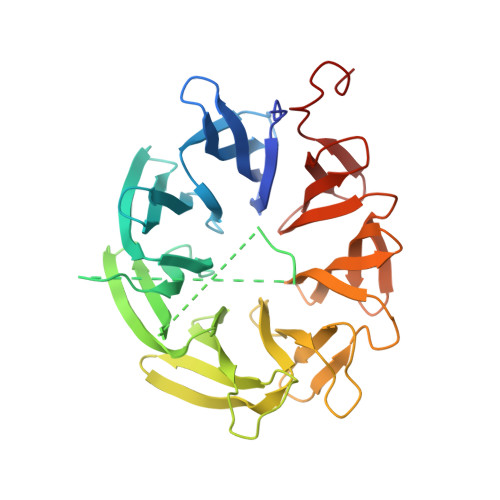
The beta-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology.
Bosch, J., Tamura, T., Tamura, N., Baumeister, W., Essen, L.O.(2007) Acta Crystallogr D Biol Crystallogr 63: 179-187
- PubMed: 17242511
- DOI: https://doi.org/10.1107/S0907444906045471
- Primary Citation of Related Structures:
2GOP - PubMed Abstract:
In the proteolytic pathway of prokaryotic and eukaryotic organisms, proteins tagged for proteolysis are firstly shredded into smaller peptides by compartmentalized proteases such as the proteasome complex. Accordingly, a variety of downstream proteases have evolved to further hydrolyze these peptides to the level of free amino acids. In the search for such downstream proteases, a high-molecular-weight protease complex called trilobed protease (TLP) was recently discovered in the archaeon Pyroccocus furiosus. The crystal structure of the N-terminal beta-propeller domain of the trilobed protease at 2 A resolution shows that the trilobed protease utilizes this accessory domain to control substrate access to the active site. Modelling of the intact TLP monomer suggests that this protease has an additional side entrance to its active site as in the DPP-IV or tricorn protease complexes.
Organizational Affiliation:
Max-Planck-Institute for Biochemistry, Department of Molecular Structural Biology, Am Klopferspitz 18a, D-82152 Martinsried, Germany.