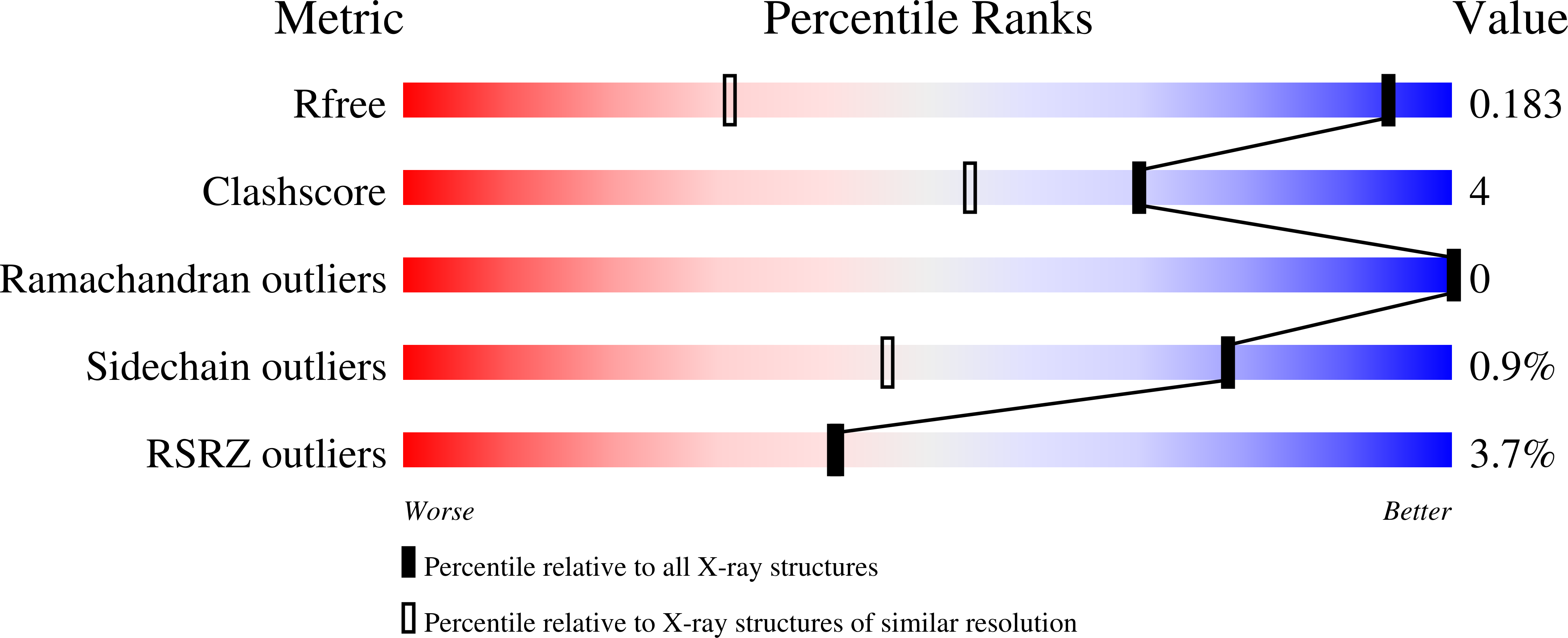
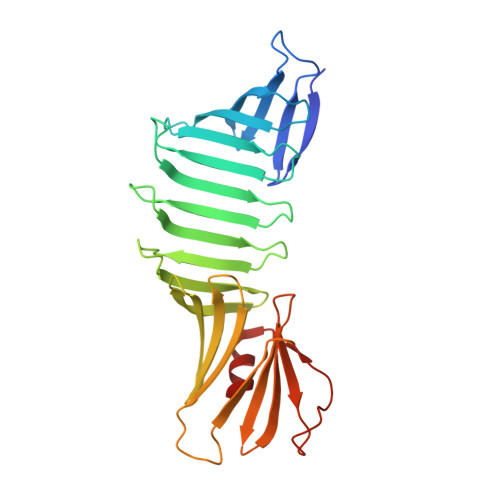
Atomic-resolution crystal structure of Borrelia burgdorferi outer surface protein A via surface engineering.
Makabe, K., Tereshko, V., Gawlak, G., Yan, S., Koide, S.(2006) Protein Sci 15: 1907-1914
- PubMed: 16823038
- DOI: https://doi.org/10.1110/ps.062246706
- Primary Citation of Related Structures:
2G8C - PubMed Abstract:
Outer surface protein A (OspA) from Borrelia burgdorferi has an unusual dumbbell-shaped structure in which two globular domains are connected with a "single-layer" beta-sheet (SLB). The protein is highly soluble, and it has been recalcitrant to crystallization. Only OspA complexes with Fab fragments have been successfully crystallized. OspA contains a large number of Lys and Glu residues, and these "high entropy" residues may disfavor crystal packing because some of them would need to be immobilized in forming a crystal lattice. We rationally designed a total of 13 surface mutations in which Lys and Glu residues were replaced with Ala or Ser. We successfully crystallized the mutant OspA without a bound Fab fragment and extended structure analysis to a 1.15 Angstroms resolution. The new high-resolution structure revealed a unique backbone hydration pattern of the SLB segment in which water molecules fill the "weak spots" on both faces of the antiparallel beta-sheet. These well-defined water molecules provide additional structural links between adjacent beta-strands, and thus they may be important for maintaining the rigidity of the SLB that inherently lacks tight packing afforded by a hydrophobic core. The structure also revealed new information on the side-chain dynamics and on a solvent-accessible cavity in the core of the C-terminal globular domain. This work demonstrates the utility of extensive surface mutation in crystallizing recalcitrant proteins and dramatically improving the resolution of crystal structures, and provides new insights into the stabilization mechanism of OspA.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Illinois 60637, USA.