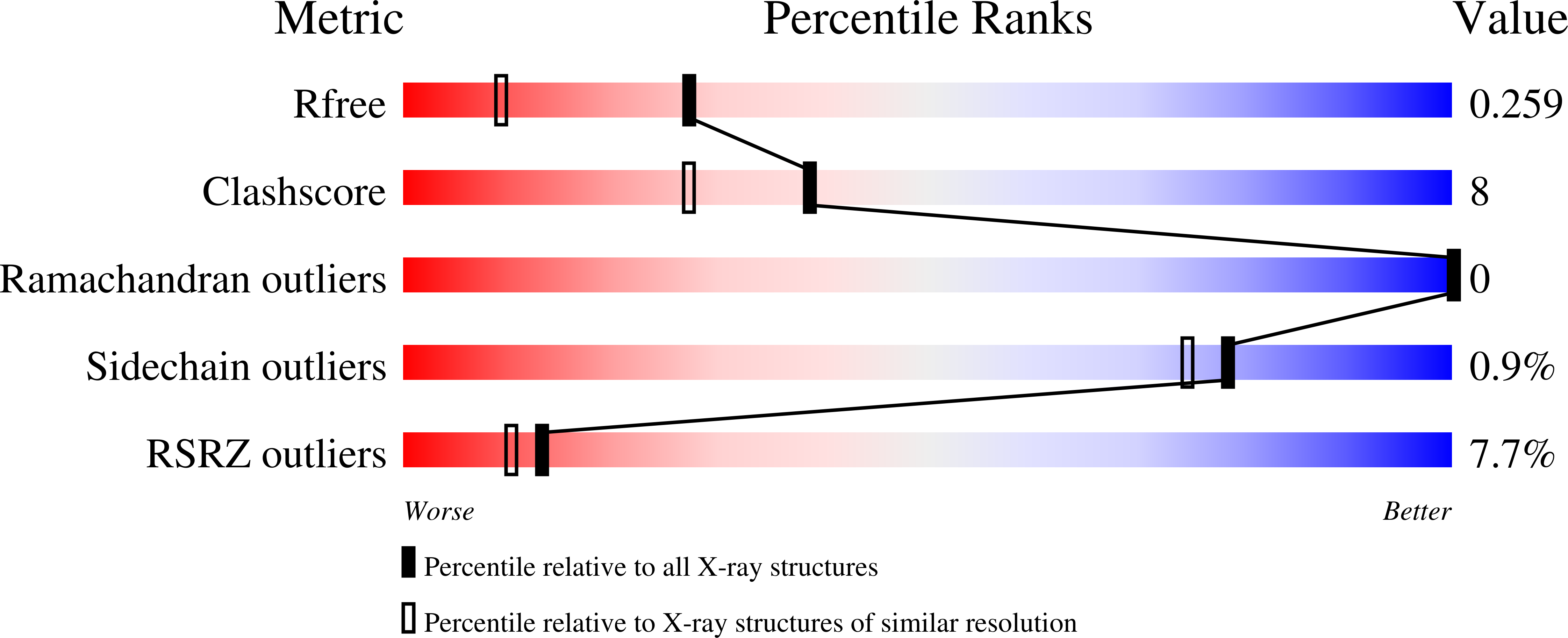
Tailoring a novel sialic acid-binding lectin from a ricin-B chain-like galactose-binding protein by natural evolution-mimicry
Yabe, R., Suzuki, R., Kuno, A., Fujimoto, Z., Jigami, Y., Hirabayashi, J.(2007) J Biochem 141: 389-399
- PubMed: 17234683
- DOI: https://doi.org/10.1093/jb/mvm043
- Primary Citation of Related Structures:
2DRY, 2DRZ, 2DS0 - PubMed Abstract:
Sialic acid (Sia) is a typical terminal sugar, which modifies various types of glycoconjugates commonly found in higher animals. Its regulatory roles in diverse biological phenomena are frequently triggered by interaction with Sia-binding lectins. When using natural Sia-binding lectins as probes, however, there have been practical problems concerning their repertoire and availability. Here, we show a rational creation of a Sia-binding lectin based on the strategy 'natural evolution-mimicry', where Sia-binding lectins are engineered by error-prone PCR from a Gal-binding lectin used as a scaffold protein. After selection with fetuin-agarose using a recently reinforced ribosome display system, one of the evolved mutants SRC showed substantial affinity for alpha2-6Sia, which the parental Gal-binding lectin EW29Ch lacked. SRC was found to have additional practical advantages in productivity and in preservation of affinity for Gal. Thus, the developed novel Sia-recognition protein will contribute as useful tools to sialoglycomics.
Organizational Affiliation:
Research Center for Glycoscience, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki 305-8568, Japan.