Structure of streptococcus pyogenes bacteriophage-associated hyaluronate lyase Hylp2
Mishra, P., Bhakuni, V., Prem Kumar, R., Singh, N., Sharma, S., Kaur, P., Singh, T.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hyaluronidase | 332 | Streptococcus pyogenes | Mutation(s): 0 EC: 4.2.2.1 | 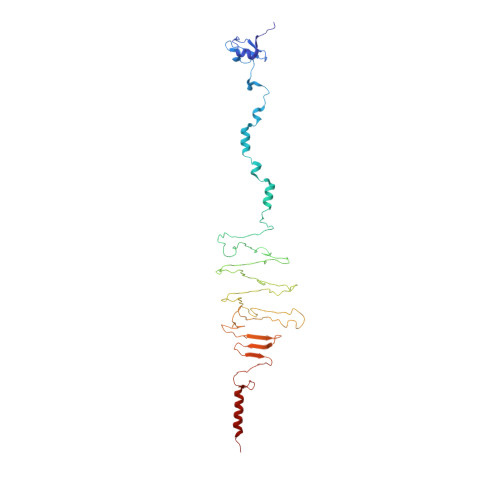 | |
UniProt | |||||
Find proteins for Q9A0M7 (Streptococcus pyogenes serotype M1) Explore Q9A0M7 Go to UniProtKB: Q9A0M7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9A0M7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.587 | α = 90 |
| b = 59.587 | β = 90 |
| c = 588.546 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |