Crystal Structure of the Human Foxk1A-DNA Complex and its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins.
Tsai, K.-L., Huang, C.-Y., Chang, C.-H., Sun, Y.-J., Chuang, W.-J., Hsiao, C.-D.(2006) J Biol Chem 281: 17400
- PubMed: 16624804
- DOI: https://doi.org/10.1074/jbc.M600478200
- Primary Citation of Related Structures:
2C6Y - PubMed Abstract:
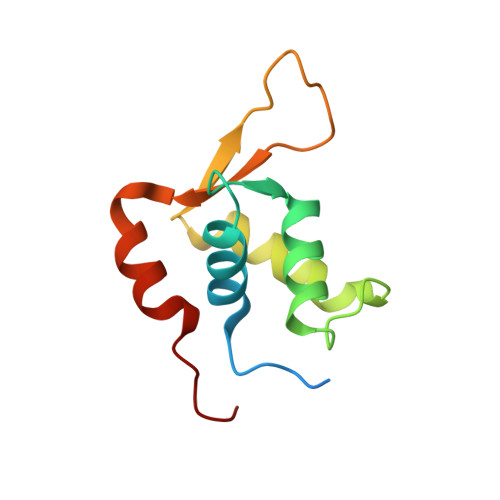
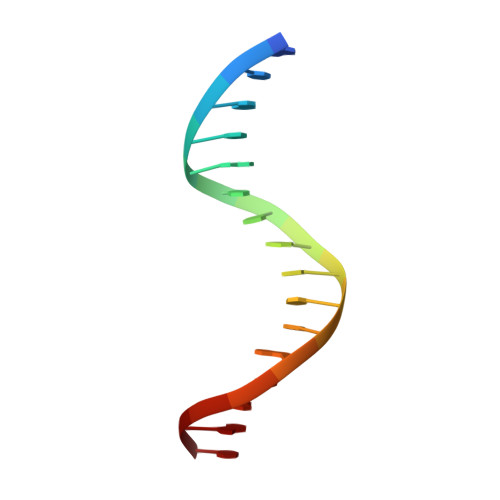
Interleukin enhancer binding factor (ILF) is a human transcription factor and a new member of the winged helix/forkhead family. ILF can bind to purine-rich regulatory motifs such as the human T-cell leukemia virus-long terminal region and the interleukin-2 promoter. Here we report the 2.4 A crystal structure of two DNA binding domains of ILF (FOXK1a) binding to a 16-bp DNA duplex containing a promoter sequence. Electrophoretic mobility shift assay studies demonstrate that two ILF-DNA binding domain molecules cooperatively bind to DNA. In addition to the recognition helix recognizing the core sequences through the major groove, the structure shows that wing 1 interacts with the minor groove of DNA, and the H2-H3 loop region makes ionic bonds to the phosphate group, which permits the recognition of DNA. The structure also reveals that the presence of the C-terminal alpha-helix in place of a typical wing 2 in a member of this family alters the orientation of the C-terminal basic residues (RKRRPR) when binding to DNA outside the core sequence. These results provide a new insight into how the DNA binding specificities of winged helix/forkhead proteins may be regulated by their less conserved regions.
Organizational Affiliation:
Institute of Molecular Biology, Academia Sinica, Taipei 115; Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu 300.