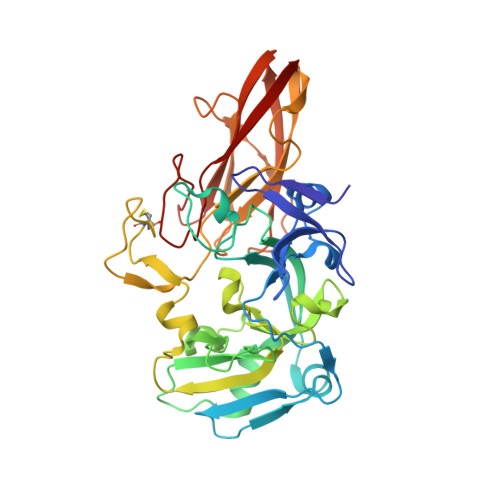
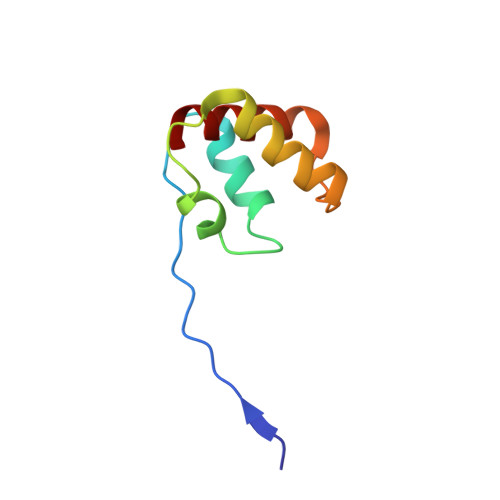
Molecular Basis of Intramolecular Electron Transfer in Sulfite-Oxidizing Enzymes is Revealed by High Resolution Structure of a Heterodimeric Complex of the Catalytic Molybdopterin Subunit and a C-Type Cytochrome Subunit.
Kappler, U., Bailey, S.(2005) J Biol Chem 280: 24999
- PubMed: 15863498
- DOI: https://doi.org/10.1074/jbc.M503237200
- Primary Citation of Related Structures:
2BLF, 2BPB - PubMed Abstract:
Sulfite-oxidizing molybdoenzymes convert the highly reactive and therefore toxic sulfite to sulfate and have been identified in insects, animals, plants, and bacteria. Although the well studied enzymes from higher animals serve to detoxify sulfite that arises from the catabolism of sulfur-containing amino acids, the bacterial enzymes have a central role in converting sulfite formed during dissimilatory oxidation of reduced sulfur compounds. Here we describe the structure of the Starkeya novella sulfite dehydrogenase, a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit, that reveals the molecular mechanism of intramolecular electron transfer in sulfite-oxidizing enzymes. The close approach of the two redox centers in the protein complex (Mo-Fe distance 16.6 A) allows for rapid electron transfer via tunnelling or aided by the protein environment. The high resolution structure of the complex has allowed the identification of potential through-bond pathways for electron transfer including a direct link via Arg-55A and/or an aromatic-mediated pathway. A potential site of electron transfer to an external acceptor cytochrome c was also identified on the SorB subunit on the opposite side to the interaction with the catalytic SorA subunit.
Organizational Affiliation:
Centre for Metals in Biology, University of Queensland, St. Lucia, Queensland 4072, Australia.