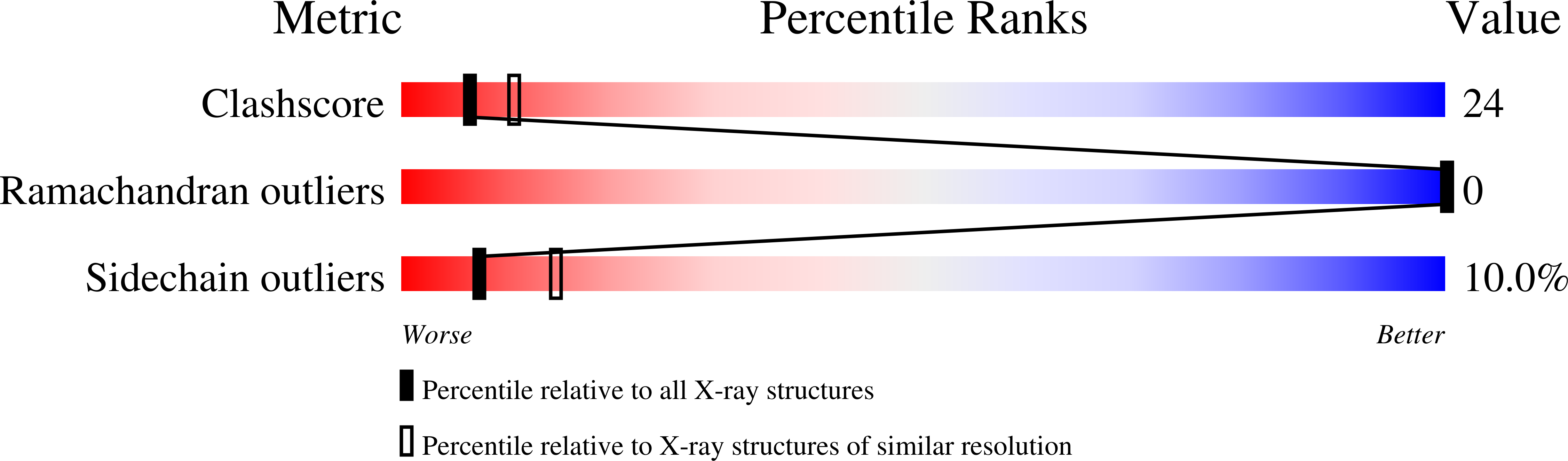Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
Ago, H., Kitagawa, Y., Fujishima, A., Matsuura, Y., Katsube, Y.(1991) J Biochem 110: 360-363
- PubMed: 1769963
- DOI: https://doi.org/10.1093/oxfordjournals.jbchem.a123586
- Primary Citation of Related Structures:
1BFG, 2BFH - PubMed Abstract:
We have determined the crystal structures of two types of human basic fibroblast growth factor, the serine analogue and the wild-type, at 1.6 and 2.5 A resolution, respectively. Two good heavy atom derivatives were found and used for multiple isomorphous replacement phasing. The atomic coordinates were refined using the Hendrickson & Konnert program for stereochemically restrained refinement against structure factors. The crystallographic R factors were reduced to 15.3% for the serine analogue structure and 16.0% for the wild-type structure. The serine analogue and wild-type structures have been found to be almost identical, the root-mean-square deviation between the corresponding C alpha atoms being 0.11 A. Their structures are composed of twelve beta-strands forming a barrel and three loops. Their molecules have an approximate threefold internal symmetry and are similar in architecture to that of interleukin-1 beta. A possible heparin-binding site, which comprises five basic residues, Lys119, Arg120, Lys125, Lys129, and Lys135, has been revealed by calculating the electrostatic potential energy.
Organizational Affiliation:
Institute for Protein Research, Osaka University.