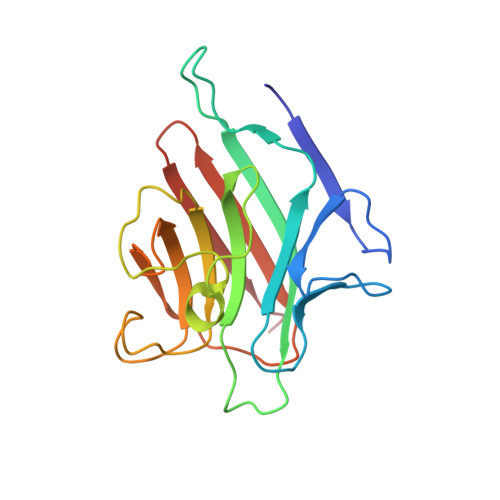
Three-dimensional structure of favin: saccharide binding-cyclic permutation in leguminous lectins
Reeke Jr., G.N., Becker, J.W.(1986) Science 234: 1108-1111
- PubMed: 3775378
- DOI: https://doi.org/10.1126/science.3775378
- Primary Citation of Related Structures:
2B7Y - PubMed Abstract:
The three-dimensional structure of favin, the glucose- and mannose-binding lectin of Vicia faba (vetch, broad bean), has been determined at a resolution of 2.8 angstroms by molecular replacement. The crystals contain specifically bound glucose and provide the first high-resolution view of specific saccharide binding in a leguminous lectin. The structure is similar to those of concanavalin A (Con A) and green pea lectin; differences from Con A show that minimal changes are needed to accommodate the cyclic permutation in amino acid sequence between the two molecules. The molecule is an ellipsoidal dimer dominated by extensive beta structures. Each protomer contains binding sites for two divalent metal ions (Mn2+ and Ca2+) and a specific saccharide. Glucose is bound by favin in a cleft in the molecular surface and has noncovalent contacts primarily with two peptide loops, one of which contains several metal ion ligands. The specific carbohydrate-binding site is similar to that of Con A in location and general peptide folding, despite several differences in specific amino acid residues.