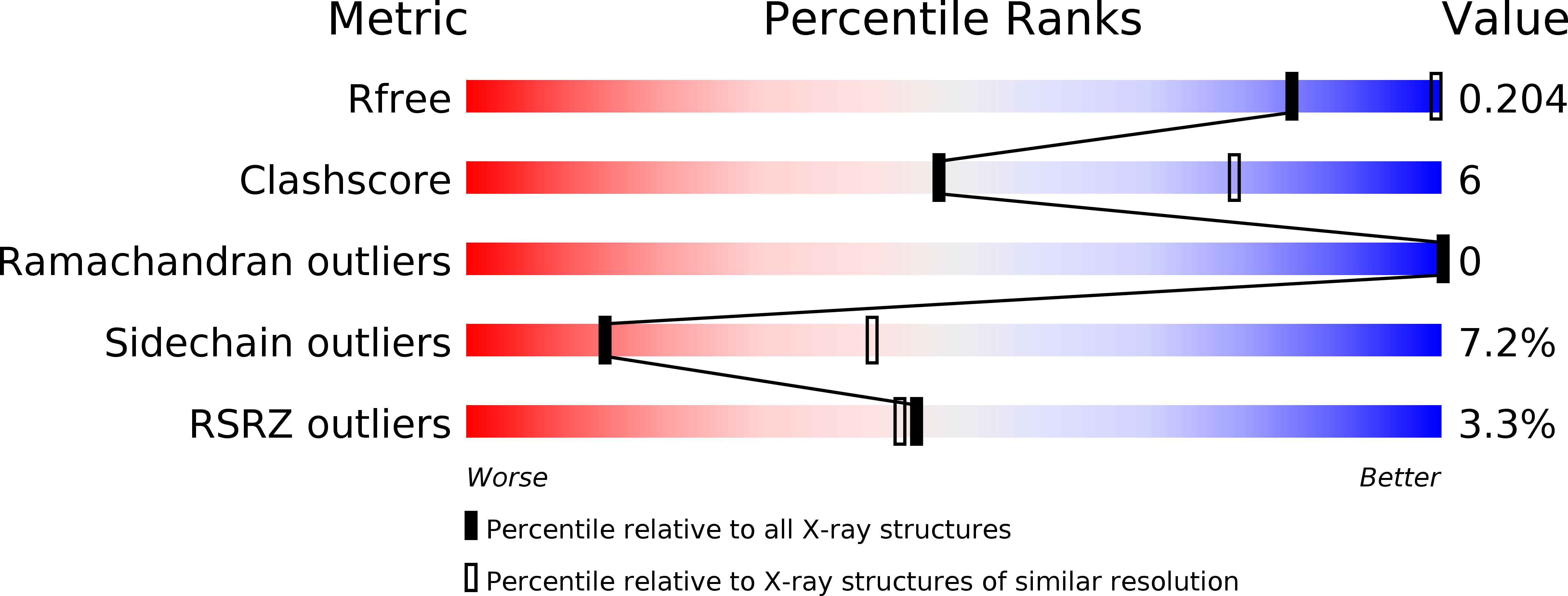
Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
Edink, E., Rucktooa, P., Retra, K., Akdemir, A., Nahar, T., Zuiderveld, O., Van Elk, R., Janssen, E., Van Nierop, P., Van Muijlwijk-Koezen, J., Smit, A.B., Sixma, T.K., Leurs, R., De Esch, I.J.P.(2011) J Am Chem Soc 133: 5363
- PubMed: 21322593
- DOI: https://doi.org/10.1021/ja110571r
- Primary Citation of Related Structures:
2Y54, 2Y56, 2Y57, 2Y58 - PubMed Abstract:
Optimization of fragment hits toward high-affinity lead compounds is a crucial aspect of fragment-based drug discovery (FBDD). In the current study, we have successfully optimized a fragment by growing into a ligand-inducible subpocket of the binding site of acetylcholine-binding protein (AChBP). This protein is a soluble homologue of the ligand binding domain (LBD) of Cys-loop receptors. The fragment optimization was monitored with X-ray structures of ligand complexes and systematic thermodynamic analyses using surface plasmon resonance (SPR) biosensor analysis and isothermal titration calorimetry (ITC). Using site-directed mutagenesis and AChBP from different species, we find that specific changes in thermodynamic binding profiles, are indicative of interactions with the ligand-inducible subpocket of AChBP. This study illustrates that thermodynamic analysis provides valuable information on ligand binding modes and is complementary to affinity data when guiding rational structure- and fragment-based discovery approaches.
Organizational Affiliation:
Leiden/Amsterdam Center of Drug Research (LACDR), Division of Medicinal Chemistry, Faculty of Sciences, VU University Amsterdam, The Netherlands.