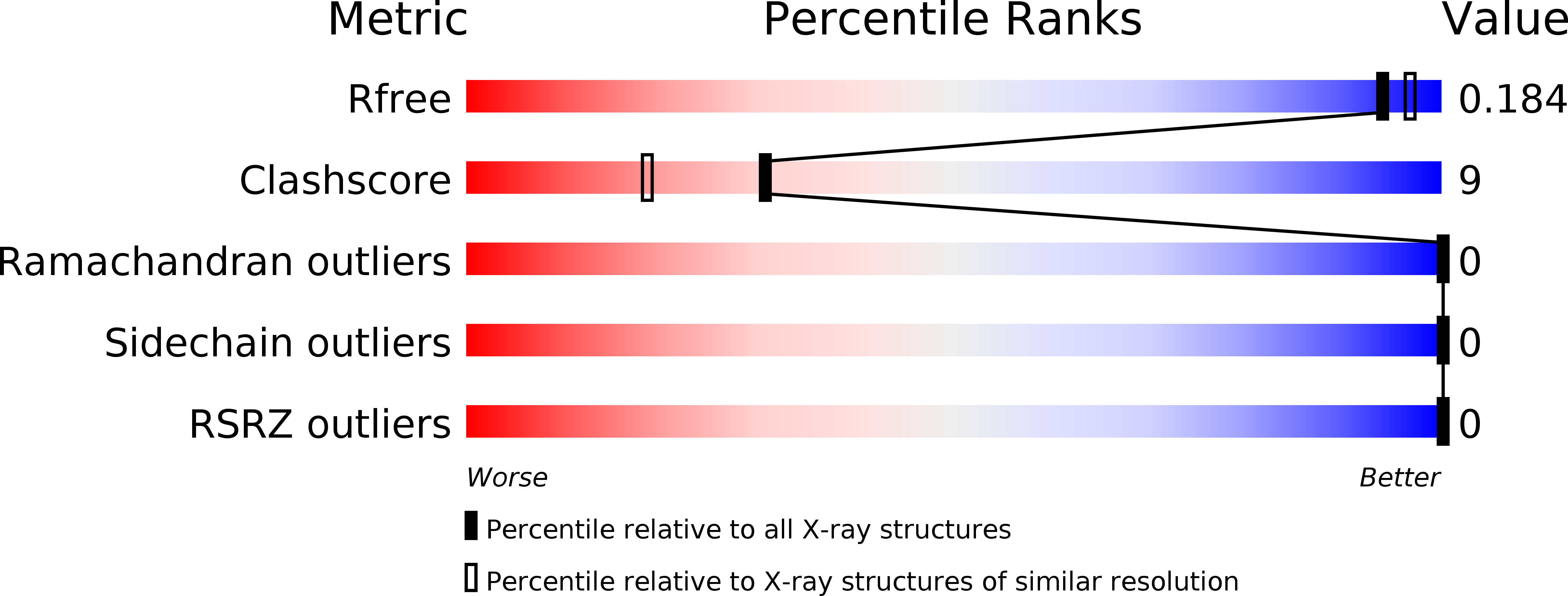
X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
Schulte, T., Hiller, R.G., Hofmann, E.(2010) FEBS Lett 584: 973
- PubMed: 20102711
- DOI: https://doi.org/10.1016/j.febslet.2010.01.041
- Primary Citation of Related Structures:
2X1Z, 2X20, 2X21 - PubMed Abstract:
The peridinin-chlorophyll a-protein (PCP) from dinoflagellates is a soluble light harvesting antenna which gathers incoming photons mainly by the carotenoid peridinin. In PCPs reconstituted with different chlorophylls, the peridinin to chlorophyll energy transfer rates are well predicted by a Förster-like theory, but only if the pigment arrangements are identical in all PCPs. We have determined the X-ray structures of PCPs reconstituted with Chlorophyll-b (Chl-b), Chlorophyll-d (Chl-d) and Bacteriochlorophyll-a (BChl-a) to resolutions
Organizational Affiliation:
Biophysics, Department of Biology and Biotechnology, Ruhr-University Bochum, D-44780 Bochum, Germany.