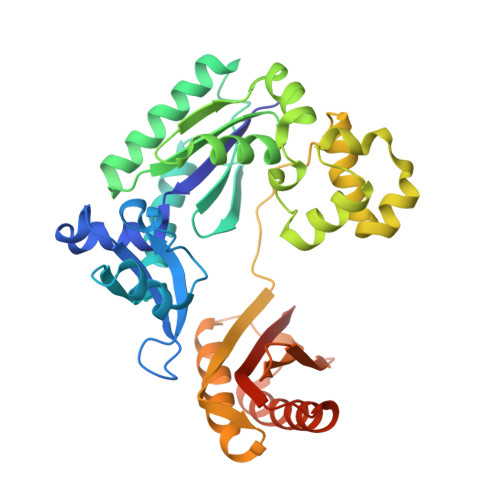
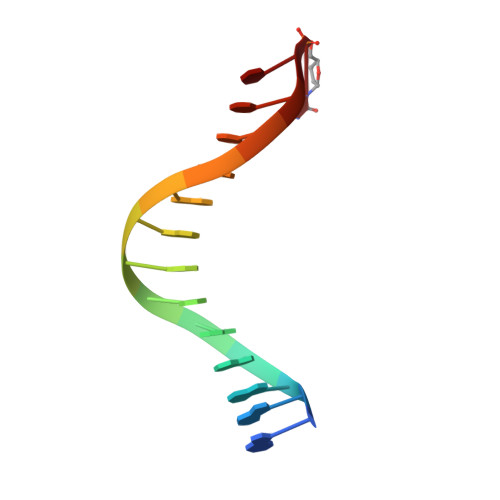
Versatility of Y-Family Sulfolobus Solfataricus DNA Polymerase Dpo4 in Translesion Synthesis Past Bulky N2-Alkylguanine Adducts.
Zhang, H., Eoff, R.L., Kozekov, I.D., Rizzo, C.J., Egli, M., Guengerich, F.P.(2009) J Biol Chem 284: 3563
- PubMed: 19059910
- DOI: https://doi.org/10.1074/jbc.M807778200
- Primary Citation of Related Structures:
2W8K, 2W8L - PubMed Abstract:
In contrast to replicative DNA polymerases, Sulfolobus solfataricus Dpo4 showed a limited decrease in catalytic efficiency (k(cat)/Km) for insertion of dCTP opposite a series of N2-alkylguanine templates of increasing size from (methyl (Me) to (9-anthracenyl)-Me (Anth)). Fidelity was maintained with increasing size up to (2-naphthyl)-Me (Naph). The catalytic efficiency increased slightly going from the N2-NaphG to the N2-AnthG substrate, at the cost of fidelity. Pre-steady-state kinetic bursts were observed for dCTP incorporation throughout the series (N2-MeG to N2-AnthG), with a decrease in the burst amplitude and k(pol), the rate of single-turnover incorporation. The pre-steady-state kinetic courses with G and all of the six N2-alkyl G adducts could be fit to a general DNA polymerase scheme to which was added an inactive complex in equilibrium with the active ternary Dpo4.DNA.dNTP complex, and only the rates of equilibrium with the inactive complex and phosphodiester bond formation were altered. Two crystal structures of Dpo4 with a template N2-NaphG (in a post-insertion register opposite a 3'-terminal C in the primer) were solved. One showed N2-NaphG in a syn conformation, with the naphthyl group located between the template and the Dpo4 "little finger" domain. The Hoogsteen face was within hydrogen bonding distance of the N4 atoms of the cytosine opposite N2-NaphG and the cytosine at the -2 position. The second structure showed N2-Naph G in an anti conformation with the primer terminus largely disordered. Collectively these results explain the versatility of Dpo4 in bypassing bulky G lesions.
Organizational Affiliation:
Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.