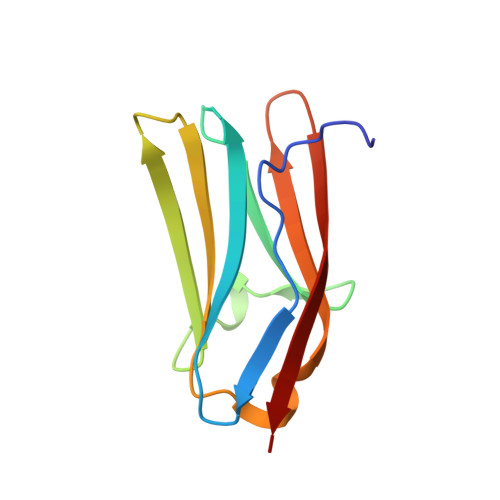
Crystal structure of the C1 domain of cardiac myosin binding protein-C: implications for hypertrophic cardiomyopathy.
Govada, L., Carpenter, L., da Fonseca, P.C., Helliwell, J.R., Rizkallah, P., Flashman, E., Chayen, N.E., Redwood, C., Squire, J.M.(2008) J Mol Biol 378: 387-397
- PubMed: 18374358
- DOI: https://doi.org/10.1016/j.jmb.2008.02.044
- Primary Citation of Related Structures:
2V6H - PubMed Abstract:
C-protein is a major component of skeletal and cardiac muscle thick filaments. Mutations in the gene encoding cardiac C-protein [cardiac myosin binding protein-C (cMyBP-C)] are one of the principal causes of hypertrophic cardiomyopathy. cMyBP-C is a string of globular domains including eight immunoglobulin-like and three fibronectin-like domains termed C0-C10. It binds to myosin and titin, and probably to actin, and may have both a structural and a regulatory role in muscle function. To help to understand the pathology of the known mutations, we have solved the structure of the immunoglobulin-like C1 domain of MyBP-C by X-ray crystallography to a resolution of 1.55 A. Mutations associated with hypertrophic cardiomyopathy are clustered at one end towards the C-terminus, close to the important C1C2 linker, where they alter the structural integrity of this region and its interactions.
Organizational Affiliation:
Biomolecular Medicine Department, SORA Division, Imperial College London, London SW7 2AZ, UK.