New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
Hillig, R.C., Eberspaecher, U.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| M-PHASE INDUCER PHOSPHATASE 2 | 203 | Homo sapiens | Mutation(s): 0 EC: 3.1.3.48 | 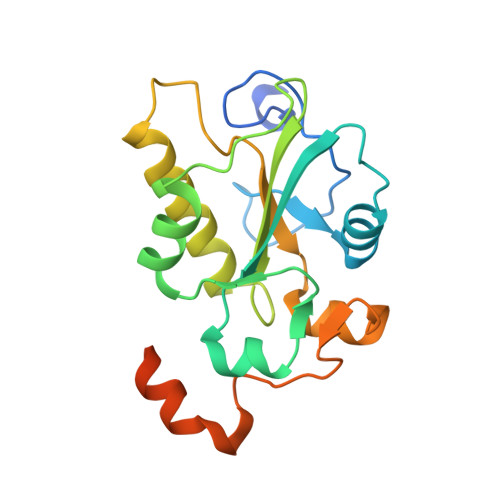 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30305 (Homo sapiens) Explore P30305 Go to UniProtKB: P30305 | |||||
PHAROS: P30305 GTEx: ENSG00000101224 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30305 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 Query on PO4 | G [auth A] H [auth B] I [auth C] J [auth D] K [auth E] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 123.925 | α = 90 |
| b = 123.925 | β = 90 |
| c = 174.035 | γ = 120 |
| Software Name | Purpose |
|---|---|
| CNX | refinement |
| MOLREP | phasing |