Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Jenkins, A.L., Zhang, Y., Ealick, S.E., Begley, T.P.(2008) Bioorg Chem 36: 29-32
- PubMed: 18054064
- DOI: https://doi.org/10.1016/j.bioorg.2007.10.005
- Primary Citation of Related Structures:
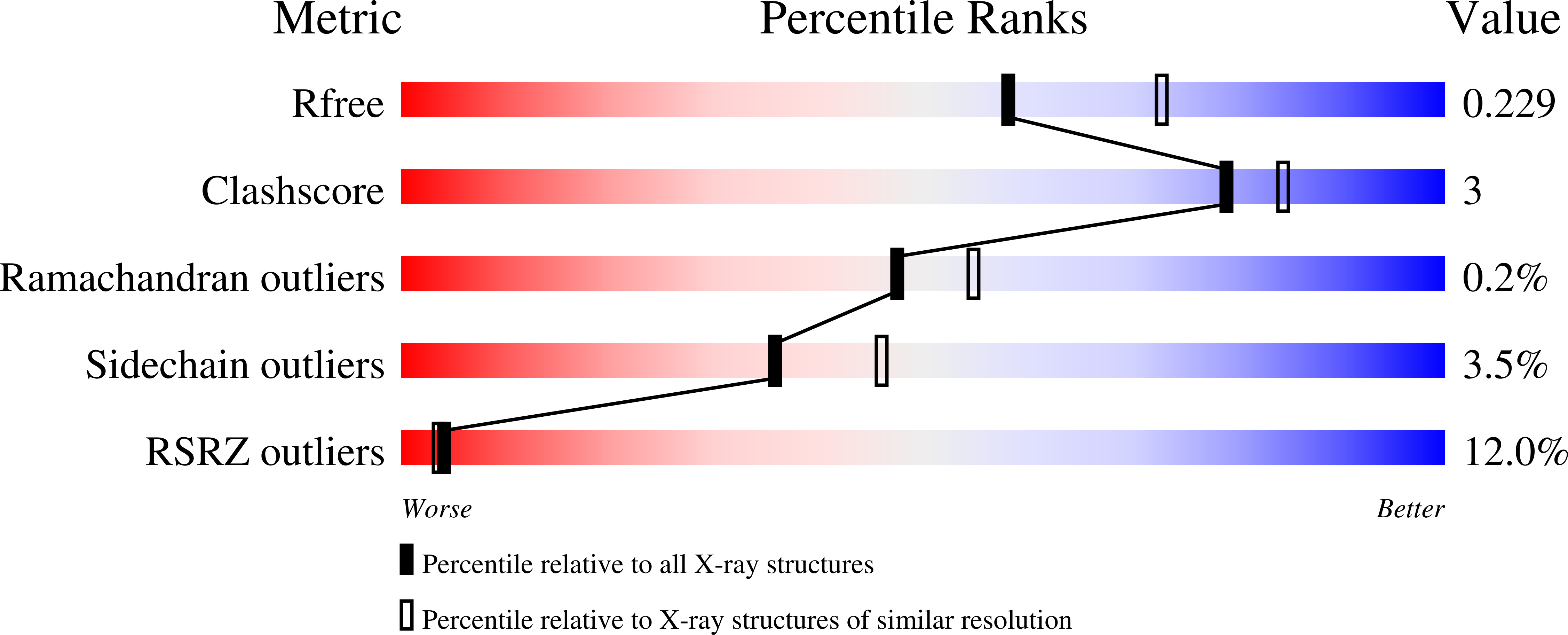
2QCX - PubMed Abstract:
TenA catalyzes the hydrolysis of 4-amino-5-aminomethyl-2-methylpyrimidine and participates in the salvage of base-degraded thiamin. Here, we describe mutagenesis of the active site of TenA guided by structures of the enzyme complexed to a substrate analog and to the product. Catalytic roles for each of the active site residues are identified and a mechanism for the reaction is described.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.