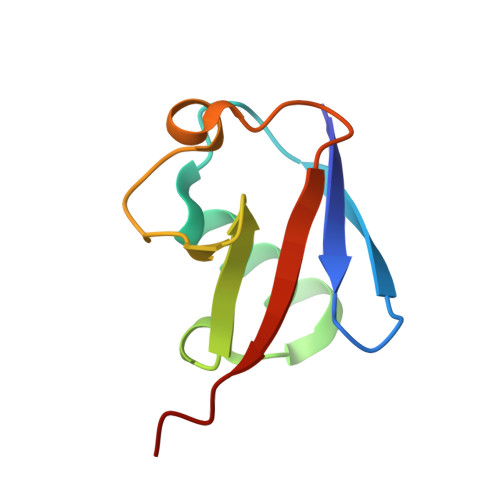
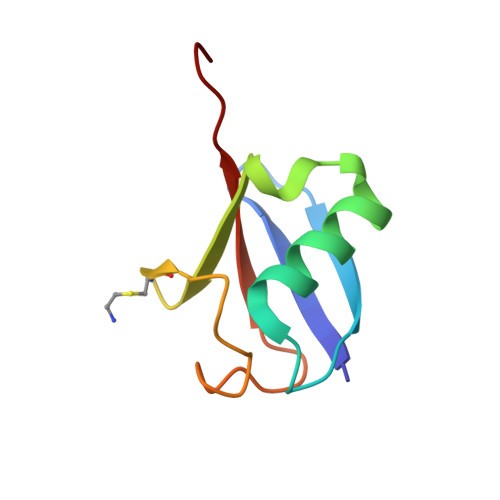
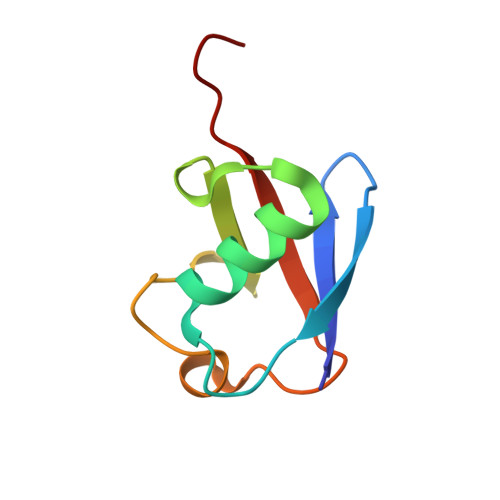
Crystal Structure and Solution NMR Studies of Lys48-linked Tetraubiquitin at Neutral pH
Eddins, M.J., Varadan, R., Fushman, D., Pickart, C.M., Wolberger, C.(2007) J Mol Biol 367: 204-211
- PubMed: 17240395
- DOI: https://doi.org/10.1016/j.jmb.2006.12.065
- Primary Citation of Related Structures:
2O6V - PubMed Abstract:
Ubiquitin modification of proteins is used as a signal in many cellular processes. Lysine side-chains can be modified by a single ubiquitin or by a polyubiquitin chain, which is defined by an isopeptide bond between the C terminus of one ubiquitin and a specific lysine in a neighboring ubiquitin. Polyubiquitin conformations that result from different lysine linkages presumably differentiate their roles and ability to bind specific targets and enzymes. However, conflicting results have been obtained regarding the precise conformation of Lys48-linked tetraubiquitin. We report the crystal structure of Lys48-linked tetraubiquitin at near-neutral pH. The two tetraubiquitin complexes in the asymmetric unit show the complete connectivity of the chain and the molecular details of the interactions. This tetraubiquitin conformation is consistent with our NMR data as well as with previous studies of diubiquitin and tetraubiquitin in solution at neutral pH. The structure provides a basis for understanding Lys48-linked polyubiquitin recognition under physiological conditions.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry and the Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.