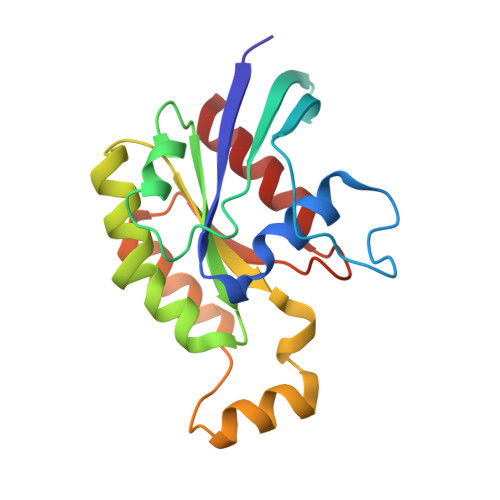
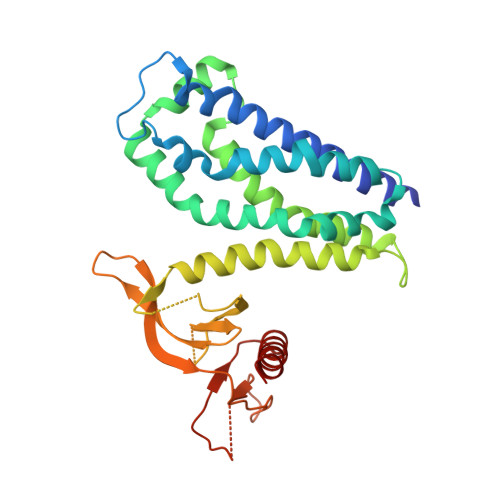
The DH and PH Domains of Trio Coordinately Engage Rho GTPases for their Efficient Activation
Chhatriwala, M.K., Betts, L., Worthylake, D.K., Sondek, J.(2007) J Mol Biol 368: 1307-1320
- PubMed: 17391702
- DOI: https://doi.org/10.1016/j.jmb.2007.02.060
- Primary Citation of Related Structures:
2NZ8 - PubMed Abstract:
Rho-family GTPases are activated by the exchange of bound GDP for GTP, a process that is catalyzed by Dbl-family guanine nucleotide exchange factors (GEFs). The catalytic unit of Dbl-family GEFs consists of a Dbl homology (DH) domain followed almost invariantly by a pleckstrin-homology (PH) domain. The majority of the catalytic interface forms between the switch regions of the GTPase and the DH domain, but full catalytic activity often requires the associated PH domain. Although PH domains are usually characterized as lipid-binding regions, they also participate in protein-protein interactions. For example, the DH-associated PH domain of Dbs must contact its cognate GTPases for efficient exchange. Similarly, the N-terminal DH/PH fragment of Trio, which catalyzes exchange on both Rac1 and RhoG, is fourfold more active in vitro than the isolated DH domain. Given continued uncertainty regarding functional roles of DH-associated PH domains, we have undertaken structural and functional analyses of the N-terminal DH/PH cassette of Trio. The crystal structure of this fragment of Trio bound to nucleotide-depleted Rac1 highlights the engagement of the PH domain with Rac1 and substitution of residues involved in this interface substantially diminishes activation of Rac1 and RhoG. Also, these mutations significantly reduce the ability of full-length Trio to induce neurite outgrowth dependent on RhoG activation in PC-12 cells. Overall, these studies substantiate a general role for DH-associated PH domains in engaging Rho GTPases directly for efficient guanine nucleotide exchange and support a parsimonious explanation for the essentially invariant linkage between DH and PH domains.
Organizational Affiliation:
Department of Pharmacology, University of North Carolina, Chapel Hill, NC 27599-7295, USA.