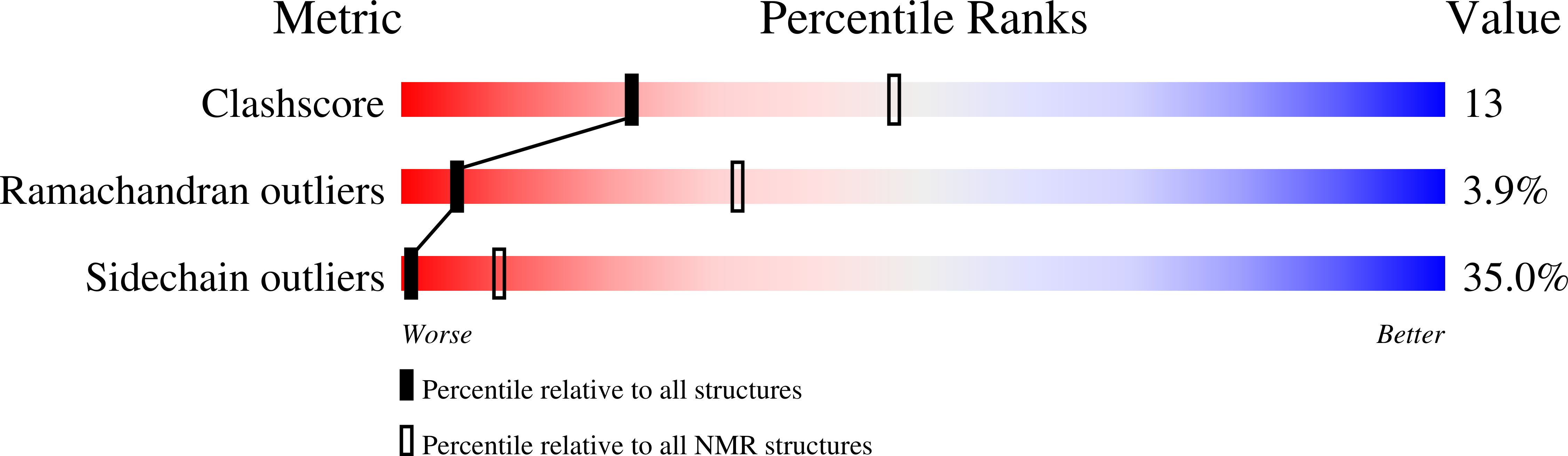Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
Datta, A., Kundu, P., Bhunia, A.(2016) J Colloid Interface Sci 461: 335-345
- PubMed: 26407061
- DOI: https://doi.org/10.1016/j.jcis.2015.09.036
- Primary Citation of Related Structures:
2N63, 2N65 - PubMed Abstract:
The remarkable rise in multi-drug resistant Gram-negative bacterial pathogens is a major concern to the well being of humans as well as susceptible plants. In recent years, diseases associated with inflammation and septicemia have already become a global health issue. Therefore, there is a rising demand for the development of novel "super" antibiotics. In this context, antimicrobial peptides offer an attractive, alternate therapeutic solution to conventional antibiotics. Microbroth dilution assay was performed to investigate the antimicrobial activities of the two designed peptides against Gram negative bacterial pathogens. Fluorescence studies including NPN dye uptake assay, Calcein entrapped vesicle leakage assay, quenching and anisotropy in presence of lipopolysaccharide (LPS) were performed to elucidate binding interactions and enhanced membrane permeabilisation. Hemolytic assay and endotoxin/LPS neutralisation assay were performed to study the hemolytic effects and LPS scavenging abilities of the peptides. High resolution NMR studies were performed to obtain insights into LPS-peptide interaction at the molecular level. Here, we report more potent analogues of previously designed peptide VG16KRKP, designed through dimerization via Cys-Cys disulphide linkage and N-terminal lipidation. Similar to the parent peptide, VG16KRKP, the modified analogue peptides are non hemolytic in nature, but possessed, 2-10-fold increase in antibacterial activities against E. coli, human pathogen Pseudomonas aeruginosa and the devastating plant pathogen, Xanthomonas campestris pv. campestris as well as membrane permeabilization, and endotoxin neutralization. LPS bound solution structure of both analogues, as determined by NMR spectroscopy, reveal that the conserved hydrophobic triad motif, formed by Trp5, Leu11 and Phe12 is compactly organized and stabilized either by the acyl chain or disulphide bond. This structural constraint accounts for the separation of polar face from the hydrophobic face of the peptides. Our novel peptides designed through Cys-Cys dimerization and N-terminal lipidation, will serve as a template to develop more potent antimicrobials in future, to control plant and human diseases.
Organizational Affiliation:
Department of Biophysics, Bose Institute, P-1/12 CIT Scheme VII (M), Kolkata 700 054, India.