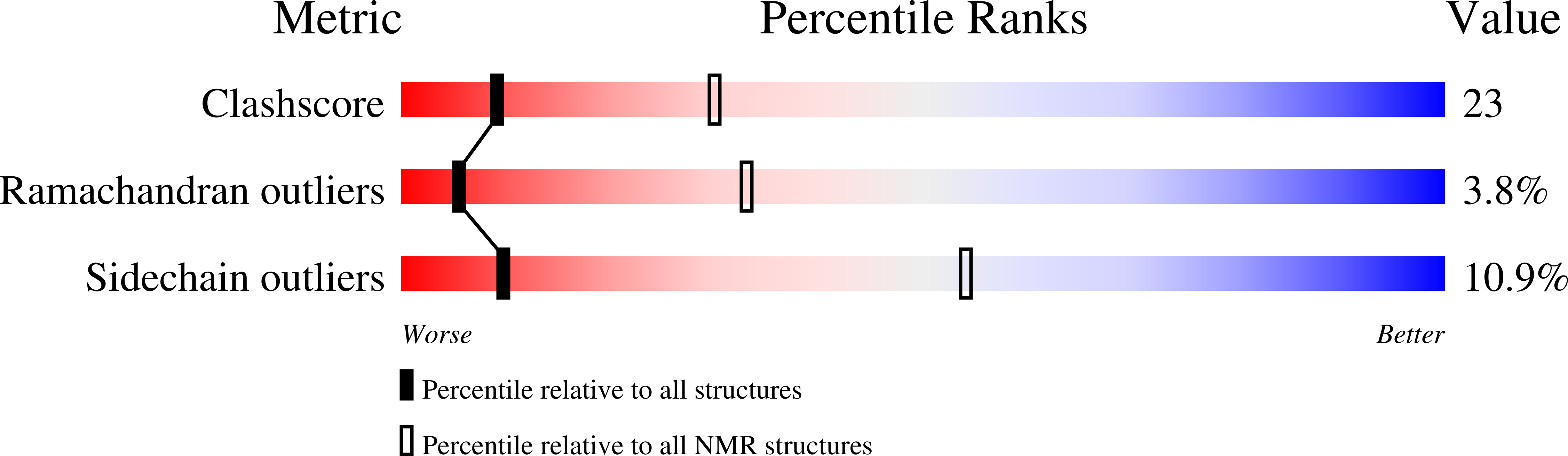The Structure of the Mercury Transporter MerF in Phospholipid Bilayers: A Large Conformational Rearrangement Results from N-Terminal Truncation.
Lu, G.J., Tian, Y., Vora, N., Marassi, F.M., Opella, S.J.(2013) J Am Chem Soc 135: 9299-9302
- PubMed: 23763519
- DOI: https://doi.org/10.1021/ja4042115
- Primary Citation of Related Structures:
2M67 - PubMed Abstract:
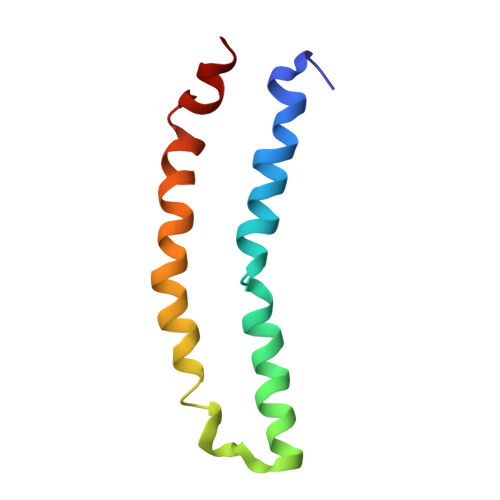
The three-dimensional structure of the 81-residue mercury transporter MerF determined in liquid crystalline phospholipid bilayers under physiological conditions by Rotationally Aligned (RA) solid-state NMR has two long helices, which extend well beyond the bilayer, with a well-defined interhelical loop. Truncation of the N-terminal 12 residues, which are mobile and unstructured when the protein is solubilized in micelles, results in a large structural rearrangement of the protein in bilayers. In the full-length protein, the N-terminal helix is aligned nearly parallel to the membrane normal and forms an extension of the first transmembrane helix. By contrast, this helix adopts a perpendicular orientation in the truncated protein. The close spatial proximity of the two Cys-containing metal binding sites in the three-dimensional structure of full-length MerF provides insights into possible transport mechanisms. These results demonstrate that major changes in protein structure can result from differences in amino acid sequence (e.g., full-length vs truncated proteins) as well as the use of a non-native membrane mimetic environment (e.g., micelles) vs liquid crystalline phospholipid bilayers. They provide further evidence of the importance of studying unmodified membrane proteins in near-native bilayer environments in order to obtain accurate structures that can be related to their functions.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093-0307, United States.