Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein.
Blondot, M.L., Dubosclard, V., Fix, J., Lassoued, S., Aumont-Nicaise, M., Bontems, F., Eleouet, J.F., Sizun, C.(2012) PLoS Pathog 8: e1002734-e1002734
- PubMed: 22675274
- DOI: https://doi.org/10.1371/journal.ppat.1002734
- Primary Citation of Related Structures:
2L9J - PubMed Abstract:
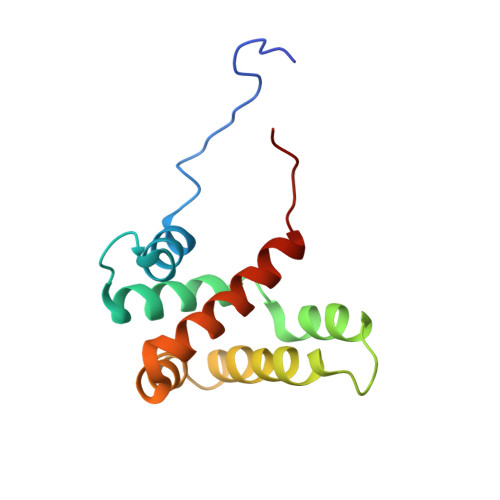
Respiratory syncytial virus (RSV) protein M2-1 functions as an essential transcriptional cofactor of the viral RNA-dependent RNA polymerase (RdRp) complex by increasing polymerase processivity. M2-1 is a modular RNA binding protein that also interacts with the viral phosphoprotein P, another component of the RdRp complex. These binding properties are related to the core region of M2-1 encompassing residues S58 to K177. Here we report the NMR structure of the RSV M2-1(58-177) core domain, which is structurally homologous to the C-terminal domain of Ebola virus VP30, a transcription co-factor sharing functional similarity with M2-1. The partial overlap of RNA and P interaction surfaces on M2-1(58-177), as determined by NMR, rationalizes the previously observed competitive behavior of RNA versus P. Using site-directed mutagenesis, we identified eight residues located on these surfaces that are critical for an efficient transcription activity of the RdRp complex. Single mutations of these residues disrupted specifically either P or RNA binding to M2-1 in vitro. M2-1 recruitment to cytoplasmic inclusion bodies, which are regarded as sites of viral RNA synthesis, was impaired by mutations affecting only binding to P, but not to RNA, suggesting that M2-1 is associated to the holonucleocapsid by interacting with P. These results reveal that RNA and P binding to M2-1 can be uncoupled and that both are critical for the transcriptional antitermination function of M2-1.
Organizational Affiliation:
Unité de Virologie et Immunologie Moléculaires (UR892), INRA, Jouy-en-Josas, France.