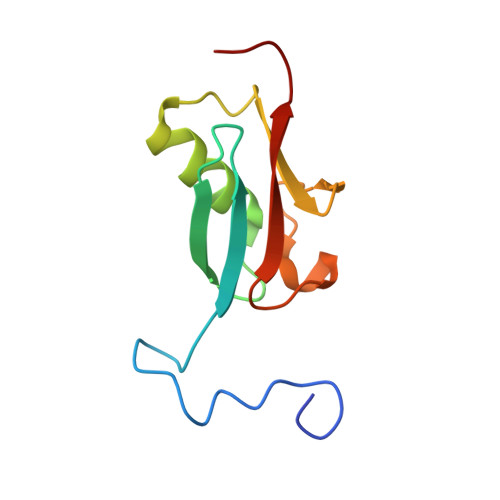
Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation.
Chang, C.C., Naik, M.T., Huang, Y.S., Jeng, J.C., Liao, P.H., Kuo, H.Y., Ho, C.C., Hsieh, Y.L., Lin, C.H., Huang, N.J., Naik, N.M., Kung, C.C., Lin, S.Y., Chen, R.H., Chang, K.S., Huang, T.H., Shih, H.M.(2011) Mol Cell 42: 62-74
- PubMed: 21474068
- DOI: https://doi.org/10.1016/j.molcel.2011.02.022
- Primary Citation of Related Structures:
2KQS - PubMed Abstract:
Small ubiquitin-like modifier (SUMO) conjugation and interaction are increasingly associated with various cellular processes. However, little is known about the cellular signaling mechanisms that regulate proteins for distinct SUMO paralog conjugation and interactions. Using the transcriptional coregulator Daxx as a model, we show that SUMO paralog-selective binding and conjugation are regulated by phosphorylation of the Daxx SUMO-interacting motif (SIM). NMR structural studies show that Daxx (732)E-I-I-V-L-S-D-S-D(740) is a bona fide SIM that binds to SUMO-1 in a parallel orientation. Daxx-SIM is phosphorylated by CK2 kinase at residues S737 and S739. Phosphorylation promotes Daxx-SIM binding affinity toward SUMO-1 over SUMO-2/3, causing Daxx preference for SUMO-1 conjugation and interaction with SUMO-1-modified factors. Furthermore, Daxx-SIM phosphorylation enhances Daxx to sensitize stress-induced cell apoptosis via antiapoptotic gene repression. Our findings provide structural insights into the Daxx-SIM:SUMO-1 complex, a model of SIM phosphorylation-enhanced SUMO paralog-selective modification and interaction, and phosphorylation-regulated Daxx function in apoptosis.
Organizational Affiliation:
Institute of Biomedical Sciences, Academia Sinica, Taipei 11529, Taiwan.