Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
Cantini, F., Veggi, D., Dragonetti, S., Savino, S., Scarselli, M., Romagnoli, G., Pizza, M., Banci, L., Rappuoli, R.(2009) J Biol Chem 284: 9022-9026
- PubMed: 19196709
- DOI: https://doi.org/10.1074/jbc.C800214200
- Primary Citation of Related Structures:
2KC0 - PubMed Abstract:
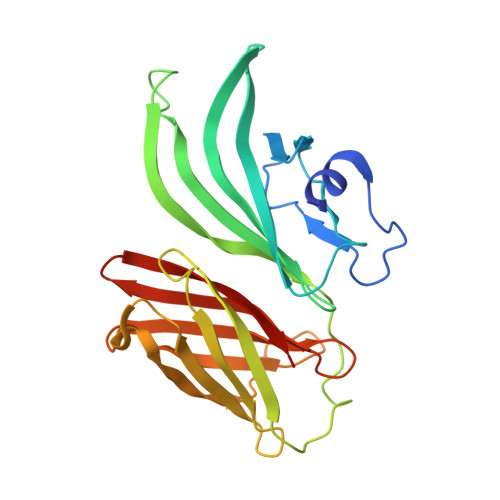
Factor H-binding protein is a 27-kDa lipoprotein of Neisseria meningitidis discovered while screening the bacterial genome for vaccine candidates. In addition to being an important component of a vaccine against meningococcus in late stage of development, the protein is essential for pathogenesis because it allows the bacterium to survive and grow in human blood by binding the human complement factor H. We recently reported the solution structure of the C-terminal domain of factor H-binding protein, which contains the immunodominant epitopes. In the present study, we report the structure of the full-length molecule, determined by nuclear magnetic resonance spectroscopy. The protein is composed of two independent barrels connected by a short link. Mapping the residues recognized by monoclonal antibodies with bactericidal or factor H binding inhibition properties allowed us to predict the sites involved in the function of the protein. The structure therefore provides the basis for designing improved vaccine molecules.
Organizational Affiliation:
Magnetic Resonance Center (CERM), F50019 Sesto Fiorentino, Italy.