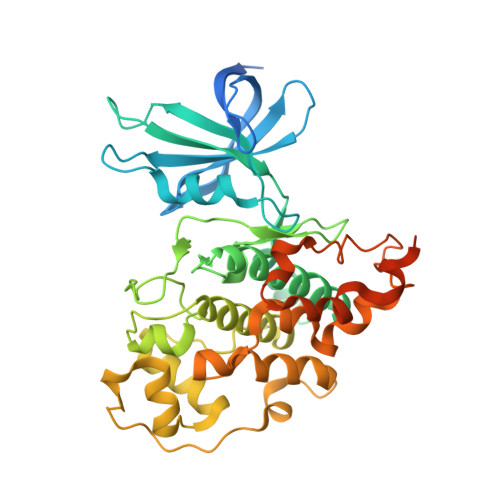
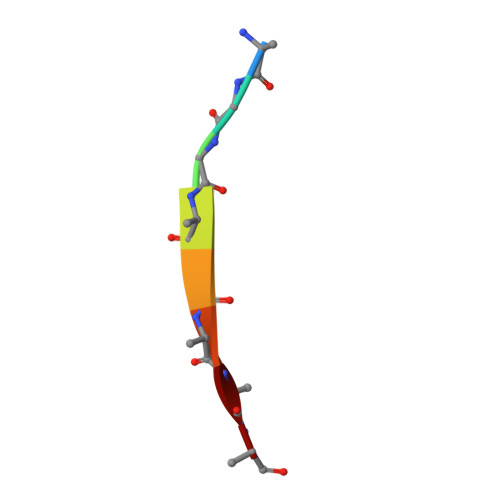
Extremely Tight Binding of a Ruthenium Complex to Glycogen Synthase Kinase 3.
Atilla-Gokcumen, G.E., Pagano, N., Streu, C., Maksimoska, J., Filippakopoulos, P., Knapp, S., Meggers, E.(2008) Chembiochem 9: 2933
- PubMed: 19035373
- DOI: https://doi.org/10.1002/cbic.200800489
- Primary Citation of Related Structures:
2JLD
Organizational Affiliation:
Fachbereich Chemie, Philipps-Universität Marburg, Hans-Meerwein-Strasse, Marburg, Germany.