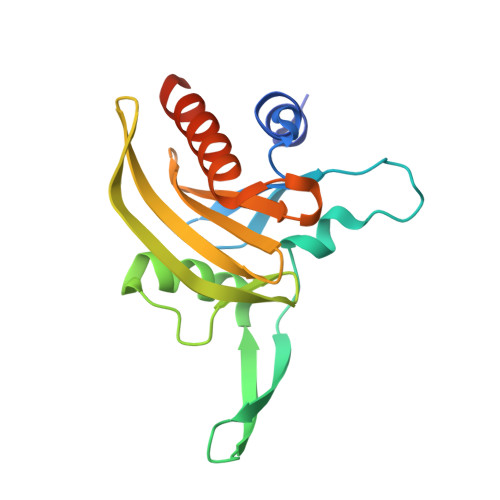
Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum.
Kursula, I., Kursula, P., Ganter, M., Panjikar, S., Matuschewski, K., Schuler, H.(2008) Structure 16: 1638
- PubMed: 19000816
- DOI: https://doi.org/10.1016/j.str.2008.09.008
- Primary Citation of Related Structures:
2JKF, 2JKG - PubMed Abstract:
Profilins are key regulators of actin dynamics. They sequester actin monomers, forming a pool for rapid polymer formation stimulated by proteins such as formins. Apicomplexan parasites utilize a highly specialized microfilament system for motility and host cell invasion. Their genomes encode only a small number of divergent actin regulators. We present the first crystal structure of an apicomplexan profilin, that of the malaria parasite Plasmodium falciparum, alone and in complex with a polyproline ligand peptide. The most striking feature of Plasmodium profilin is a unique minidomain consisting of a large beta-hairpin extension common to all apicomplexan parasites, and an acidic loop specific for Plasmodium species. Reverse genetics in the rodent malaria model, Plasmodium berghei, suggests that profilin is essential for the invasive blood stages of the parasite. Together, our data establish the structural basis for understanding the functions of profilin in the malaria parasite.
Organizational Affiliation:
Department of Biochemistry, University of Oulu, 90570 Oulu, Finland. inari.kursula@oulu.fi