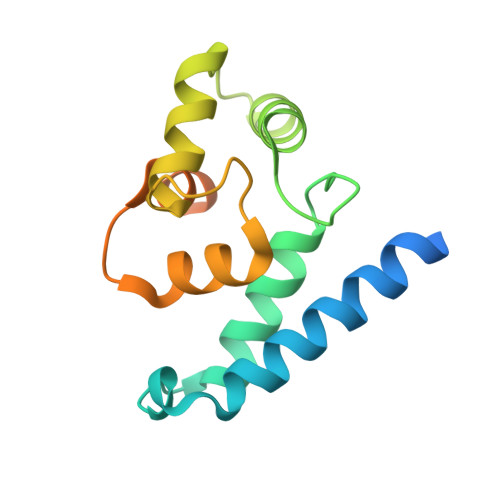
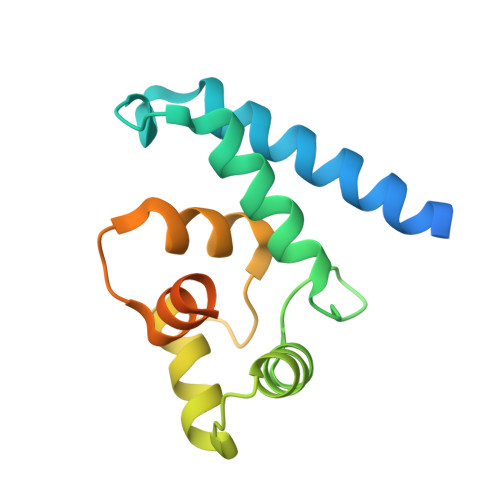
Structural and Functional Characterization of Human Iba Proteins.
Schulze, J.O., Quedenau, C., Roske, Y., Adam, T., Schuler, H., Behlke, J., Turnbull, A.P., Sievert, V., Scheich, C., Mueller, U., Heinemann, U., Bussow, K.(2008) FEBS J 275: 4627
- PubMed: 18699778
- DOI: https://doi.org/10.1111/j.1742-4658.2008.06605.x
- Primary Citation of Related Structures:
2JJZ, 2VTG - PubMed Abstract:
Iba2 is a homolog of ionized calcium-binding adapter molecule 1 (Iba1), a 17-kDa protein that binds and cross-links filamentous actin (F-actin) and localizes to membrane ruffles and phagocytic cups. Here, we present the crystal structure of human Iba2 and its homodimerization properties, F-actin cross-linking activity, cellular localization and recruitment upon bacterial invasion in comparison with Iba1. The Iba2 structure comprises two central EF-hand motifs lacking bound Ca2+. Iba2 crystallized as a homodimer stabilized by a disulfide bridge and zinc ions. Analytical ultracentrifugation revealed a different mode of dimerization under reducing conditions that was independent of Ca2+. Furthermore, no binding of Ca2+ up to 0.1 mM was detected by equilibrium dialysis. Correspondingly, Iba EF-hand motifs lack residues essential for strong Ca2+ coordination. Sedimentation experiments and microscopy detected pronounced, indistinguishable F-actin binding and cross-linking activity of Iba1 and Iba2 with induction of F-actin bundles. Fluorescent Iba fusion proteins were expressed in HeLa cells and co-localized with F-actin. Iba1 was recruited into cellular projections to a larger extent than Iba2. Additionally, we studied Iba recruitment in a Shigella invasion model that induces cytoskeletal rearrangements. Both proteins were recruited into the bacterial invasion zone and Iba1 was again concentrated slightly higher in the cellular extensions.
Organizational Affiliation:
Max Delbrück Center for Molecular Medicine, Berlin, Germany.