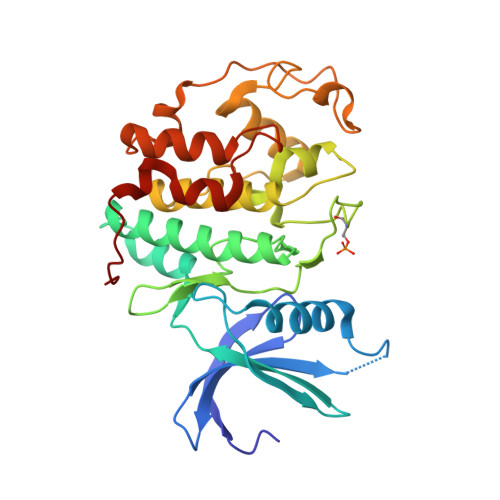
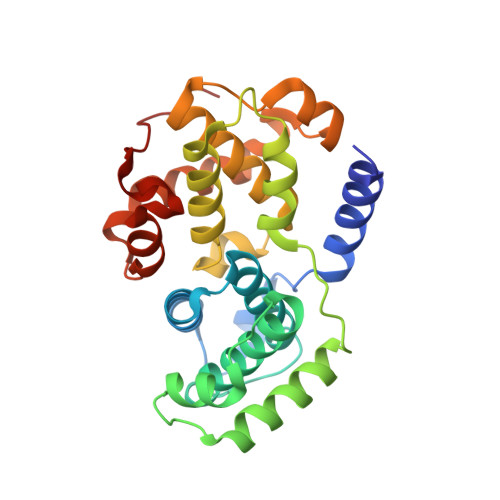
Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
Pratt, D.J., Bentley, J., Jewsbury, P., Boyle, F.T., Endicott, J.A., Noble, M.E.M.(2006) J Med Chem 49: 5470
- PubMed: 16942020
- DOI: https://doi.org/10.1021/jm060216x
- Primary Citation of Related Structures:
2IW6, 2IW8, 2IW9 - PubMed Abstract:
Cyclin dependent kinases are a key family of kinases involved in cell cycle regulation and are an attractive target for cancer chemotherapy. The roles of four residues of the cyclin-dependent kinase active site in inhibitor selectivity were investigated by producing cyclin-dependent kinase 2 mutants bearing equivalent cyclin-dependent kinase 4 residues, namely F82H, L83V, H84D, and K89T. Assay of the mutants with a cyclin-dependent kinase 4-selective bisanilinopyrimidine shows that the K89T mutation is primarily responsible for the selectivity of this compound. Use of the cyclin-dependent kinase 2-selective 6-cyclohexylmethoxy-2-(4'-sulfamoylanilino)purine (NU6102) shows that K89T has no role in the selectivity, while the remaining three mutations have a cumulative influence. The results indicate that certain residues that are not frequently considered in structure-aided kinase inhibitor design have an important role to play.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.