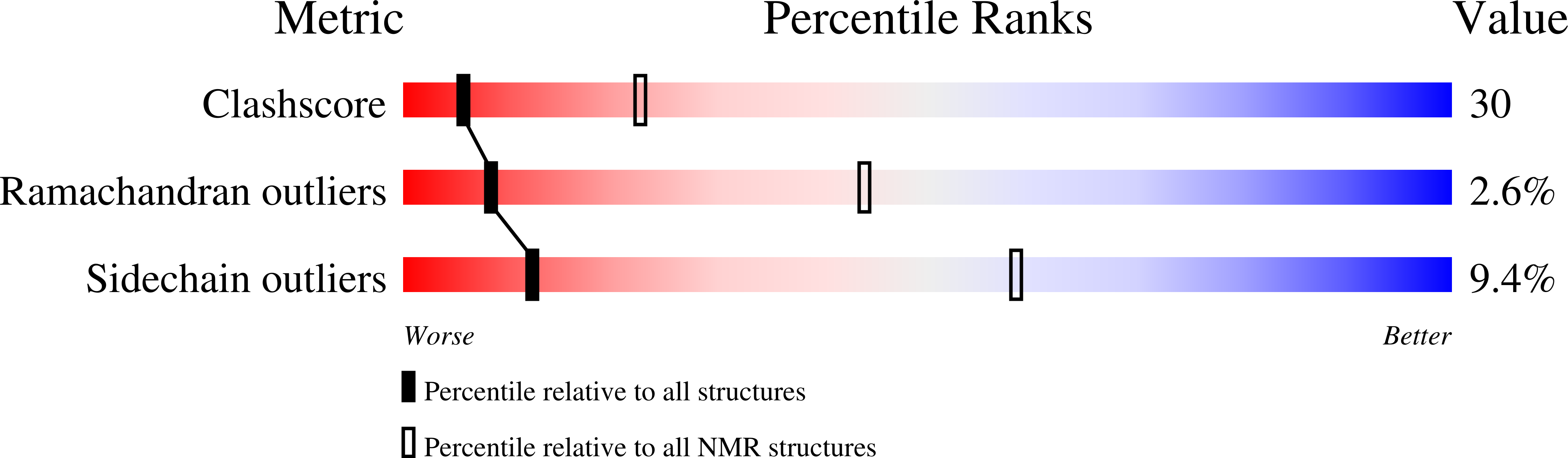The structure of a novel insect peptide explains its Ca2+ channel blocking and antifungal activities
Kouno, T., Mizuguchi, M., Tanaka, H., Yang, P., Mori, Y., Shinoda, H., Unoki, K., Aizawa, T., Demura, M., Suzuki, K., Kawano, K.(2007) Biochemistry 46: 13733-13741
- PubMed: 17994764
- DOI: https://doi.org/10.1021/bi701319t
- Primary Citation of Related Structures:
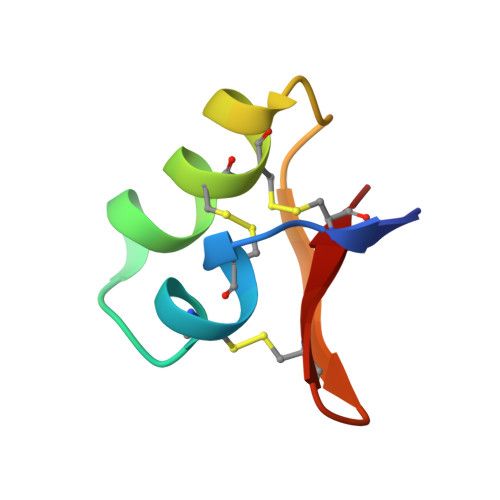
2E2F - PubMed Abstract:
Diapause-specific peptide (DSP), derived from the leaf beetle, inhibits Ca2+ channels and has antifungal activity. DSP acts on chromaffin cells of the adrenal medulla in a fashion similar to that of omega-conotoxin GVIA, a well-known neurotoxic peptide, and blocks N-type voltage-dependent Ca2+ channels. However, the amino acid sequence of DSP has little homology with any other known Ca2+ channel blockers or antifungal peptides. In this paper, we analyzed the solution structure of DSP by using two-dimensional 1H nuclear magnetic resonance and determined the pairing of half-cystine residues forming disulfide bonds. The arrangement of the three disulfide bridges in DSP was distinct from that of other antifungal peptides and conotoxins. The overall structure of DSP is compact due in part to the three disulfide bridges and, interestingly, is very similar to those of the insect- and plant-derived antifungal peptides. On the other hand, the disulfide arrangement and the three-dimensional structure of DSP and GVIA are not similar. Nevertheless, some surface residues of DSP superimpose on the key functional residues of GVIA. This homologous distribution of hydrophobic and charged side chains may result in the functional similarity between DSP and GVIA. Thus, we propose here that the three-dimensional structure of DSP can explain its dual function as a Ca2+ channel blocker and antifungal peptide.
Organizational Affiliation:
Faculty of Pharmaceutical Sciences, University of Toyama, Toyama 930-0194, Japan.