Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Roberts, E.L., Shu, N., Howard, M.J., Broadhurst, R.W., Chapman-Smith, A., Wallace, J.C., Morris, T., Cronan Jr., J.E., Perham, R.N.(1999) Biochemistry 38: 5045-5053
- PubMed: 10213607
- DOI: https://doi.org/10.1021/bi982466o
- Primary Citation of Related Structures:
2BDO, 3BDO - PubMed Abstract:
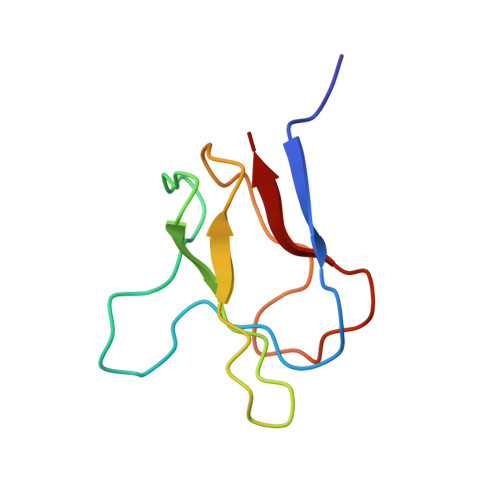
A subgene encoding the 87 C-terminal amino acids of the biotinyl carboxy carrier protein (BCCP) from the acetyl CoA carboxylase of Escherichia coli was overexpressed and the apoprotein biotinylated in vitro. The structures of both the apo and holo forms of the biotinyl domain were determined by means of multidimensional NMR spectroscopy. That of the holo domain was well-defined, except for the 10 N-terminal residues, which form part of the flexible linker between the biotinyl and subunit-binding domains of BCCP. In agreement with X-ray crystallographic studies [Athappilly, F. K., and Hendrickson, W. A. (1995) Structure 3, 1407-1419], the structure comprises a flattened beta-barrel composed of two four-stranded beta-sheets with a 2-fold axis of quasi-symmetry and the biotinyl-lysine residue displayed in an exposed beta-turn on the side of the protein opposite from the N- and C-terminal residues. The biotin group is immobilized on the protein surface, with the ureido ring held down by interactions with a protruding polypeptide "thumb" formed by residues 94-101. However, at the site of carboxylation, no evidence could be found in solution for the predicted hydrogen bond between the main chain O of Thr94 and the ureido HN1'. The structure of the apo domain is essentially identical, although the packing of side chains is more favorable in the holo domain, and this may be reflected in differences in the dynamics of the two forms. The thumb region appears to be lacking in almost all other biotinyl domain sequences, and it may be that the immobilization of the biotinyl-lysine residue in the biotinyl domain of BCCP is an unusual requirement, needed for the catalytic reaction of acetyl CoA carboxylase.
Organizational Affiliation:
Cambridge Centre for Molecular Recognition, Department of Biochemistry, University of Cambridge, U.K.