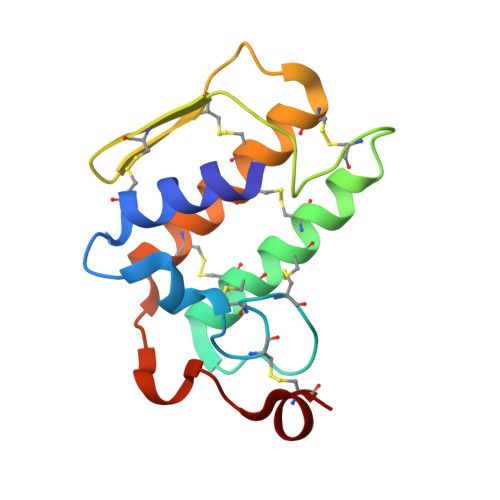
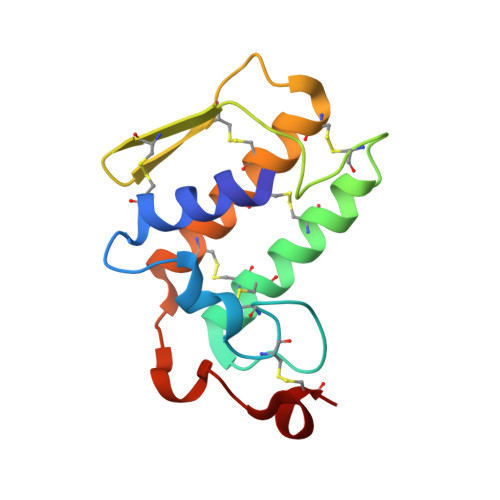
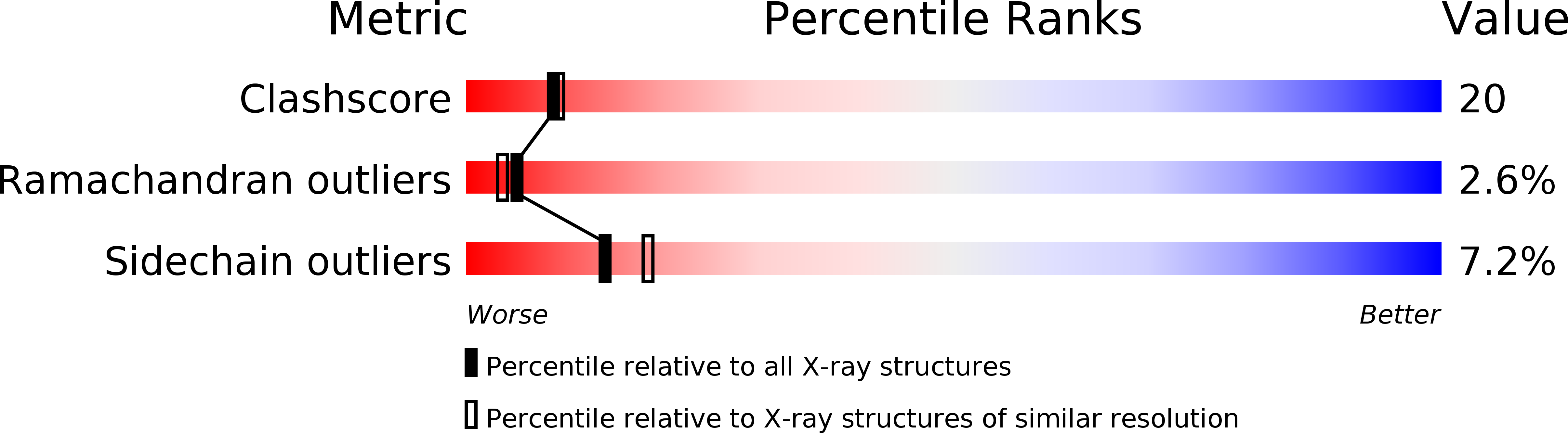Crystal structure of a heterodimer of phospholipase A2 from Naja naja sagittifera at 2.3 A resolution reveals the presence of a new PLA2-like protein with a novel cys 32-Cys 49 disulphide bridge with a bound sugar at the substrate-binding site
Jabeen, T., Singh, N., Singh, R.K., Jasti, J., Sharma, S., Kaur, P., Srinivasan, A., Singh, T.P.(2006) Proteins 62: 329-337
- PubMed: 16287060
- DOI: https://doi.org/10.1002/prot.20708
- Primary Citation of Related Structures:
1Y75 - PubMed Abstract:
The crystal structure of the phospholipase A2 (PLA2) heterodimer from Naja naja sagittifera reveals the presence of a new PLA2-like protein with eight disulphide bridges. The heterodimer is formed between a commonly observed group I PLA2 having seven characteristic disulfide bonds and a novel PLA2-like protein (Cys-PLA2) containing two extra cysteines at two highly conserved sites (positions 32 and 49) of structural and functional importance. The crystals of the heterodimer belong to tetragonal space group P41212 with cell dimensions, a = b = 77.7 A and c = 68.4 A corresponding to a solvent content of 33%, which is one of the lowest values observed so far in the PLA2 crystals. The structure has been solved with molecular replacement method and refined to a final R value of 21.6% [Rfree = 25.6%]. The electron density revealed the presence of cysteines 32 and 49 that are covalently linked to give rise to an eighth disulphide bridge in the PLA2-like monomer. A non-protein high-quality electron density was also observed at the substrate-binding site in the PLA2-like protein that has been interpreted as N-acetylglucosamine. The overall tertiary folds of the two monomers are similar having all features of PLA2-type folding. A zinc ion is detected at the interface of the heterodimer with fivefold coordination while another zinc ion was found on the surface of Cys-PLA2 with sixfold coordination. The conformations of the calcium-binding loops of both monomers are significantly different from each other as well as from those in other group I PLA2s. The N-acetylglucosamine molecule is favorably placed in the substrate-binding site of Cys-PLA2 and forms five hydrogen bonds and several van der Waals interactions with protein atoms, thus indicating a strong affinity. It also provides clue of the possible mechanism of sugar recognition by PLA2 and PLA2-like proteins. The formation of heterodimer seems to have been induced by zinc ion.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.