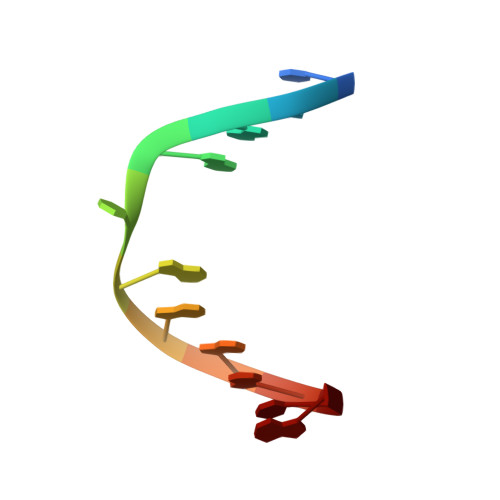
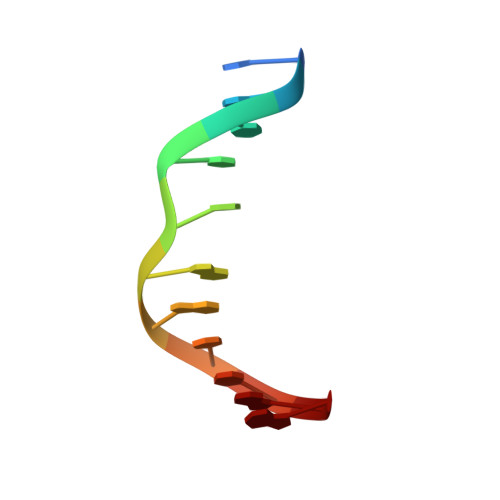
Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nakatani, K., Hagihara, S., Goto, Y., Kobori, A., Hagihara, M., Hayashi, G., Kyo, M., Nomura, M., Mishima, M., Kojima, C.(2005) Nat Chem Biol 1: 39-43
- PubMed: 16407992
- DOI: https://doi.org/10.1038/nchembio708
- Primary Citation of Related Structures:
1X26 - PubMed Abstract:
DNA trinucleotide repeats, particularly CXG, are common within the human genome. However, expansion of trinucleotide repeats is associated with a number of disorders, including Huntington disease, spinobulbar muscular atrophy and spinocerebellar ataxia. In these cases, the repeat length is known to correlate with decreased age of onset and disease severity. Repeat expansion of (CAG)n, (CTG)n and (CGG)n trinucleotides may be related to the increased stability of alternative DNA hairpin structures consisting of CXG-CXG triads with X-X mismatches. Small-molecule ligands that selectively bound to CAG repeats could provide an important probe for determining repeat length and an important tool for investigating the in vivo repeat extension mechanism. Here we report that napthyridine-azaquinolone (NA, 1) is a ligand for CAG repeats and can be used as a diagnostic tool for determining repeat length. We show by NMR spectroscopy that binding of NA to CAG repeats induces the extrusion of a cytidine nucleotide from the DNA helix.
Organizational Affiliation:
Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Kyoto 615-8510, Japan. nakatani@sanken.osaka-u.ac.jp