Crystal Structure of the NAD Biosynthetic Enzyme Quinolinate Synthase
Sakuraba, H., Tsuge, H., Yoneda, K., Katunuma, N., Ohshima, T.(2005) J Biol Chem 280: 26645-26648
- PubMed: 15937336
- DOI: https://doi.org/10.1074/jbc.C500192200
- Primary Citation of Related Structures:
1WZU - PubMed Abstract:
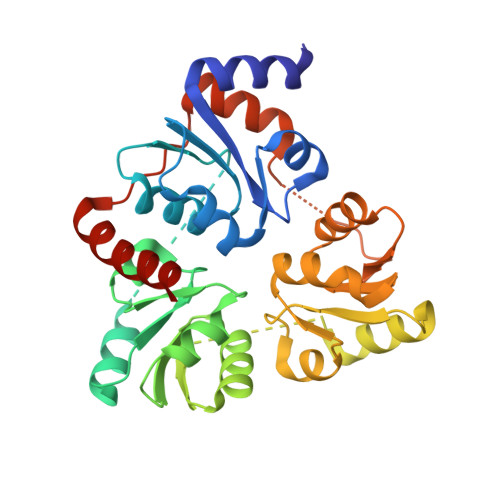
A gene encoding a quinolinate synthase has been identified in the hyperthermophilic archaeon Pyrococcus horikoshii via genome sequencing. The gene was overexpressed in Escherichia coli, and the crystal structure of the produced enzyme was determined to 2.0 A resolution in the presence of malate, a substrate analogue. The overall structure exhibits a unique triangular architecture composed of a 3-fold repeat of three-layer (alphabetaalpha) sandwich folding. Although some aspects of the fold homologous to the each domain have been observed previously, the overall structure of quinolinate synthase shows no similarity to any known protein structure. The three analogous domains are related to a pseudo-3-fold symmetry. The active site is located at the interface of the three domains and is centered on the pseudo-3-fold axis. The malate molecule is tightly held near the bottom of the active site cavity. The model of the catalytic state during the first condensation step of the quinolinate synthase reaction indicates that the elimination of inorganic phosphate from dihydroxyacetone phosphate may precede the condensation reaction.
Organizational Affiliation:
Department of Biological Science and Technology, Faculty of Engineering, The University of Tokushima, 2-1 Minamijosanjima-cho, Tokushima 770-8506.